Tankyrase polymerization is controlled by its sterile alpha motif and poly(ADP-ribose) polymerase domains - PubMed (original) (raw)
Tankyrase polymerization is controlled by its sterile alpha motif and poly(ADP-ribose) polymerase domains
Manu De Rycker et al. Mol Cell Biol. 2004 Nov.
Abstract
Tankyrases are novel poly(ADP-ribose) polymerases that have SAM and ankyrin protein-interaction domains. They are found at telomeres, centrosomes, nuclear pores, and Golgi vesicles and have been shown to participate in telomere length regulation. Their other function(s) are unknown, and it has been difficult to envision a common role at such diverse cellular locations. We have shown that tankyrase 1 polymerizes through its sterile alpha motif (SAM) domain to assemble large protein complexes. In vitro polymerization is reversible and still allows interaction with ankyrin-domain binding proteins. Polymerization can also occur in vivo, with SAM-dependent association of overexpressed tankyrase leading to formation of large tankyrase-containing vesicles, disruption of Golgi structure, and inhibition of apical secretion. Finally, tankyrase polymers are dissociated efficiently by poly(ADP-ribosy)lation. This disassembly is prevented by mutation of the PARP domain. Our findings indicate that tankyrase 1 has the unique capacity to promote both assembly and disassembly of large protein complexes. Thus, tankyrases appear to be master scaffolding proteins that regulate the formation of dynamic protein networks at different cellular locations. This implies a common scaffolding function for tankyrases at each location, with specific tankyrase interaction partners conferring location-specific roles to each network, e.g., telomere compaction or regulation of vesicle trafficking.
Figures
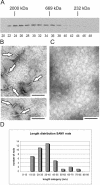
FIG. 1.
Self-association of the tankyrase 1 SAM domain. (A) Superose 6 fractionation of MBP-SAM1. Peak fractions containing molecular weight markers are indicated: 2,000 kDa, dextran blue; 669 kDa, thyroglobulin; 232 kDa, catalase. Fraction numbers are shown below. (B and C) Transmission electron microscopy of uranyl acetate-stained MBP-SAM1 polymers. Arrows indicate SAM polymers. Samples shown in panel C were treated with proteinase K prior to deposition on the EM grid. Bar, 100 nm. (D) Histogram showing the length distribution of MBP-SAM1 rods. Mean (37 nm) and median (36 nm) lengths were calculated for 40 rods, but the shortest rods were probably underrepresented because they were difficult to distinguish from background.
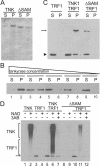
FIG. 2.
Solubility and catalytic activity of full-length tankyrase 1. (A) Solubility of recombinant tankyrase. Purified full-length tankyrase 1 (TNK) or tankyrase1-ΔSAM (ΔSAM) was centrifuged, and protein in the supernatant (S) or pellet (P) was detected by SDS-PAGE and Coomassie staining. (B) Increase in tankyrase 1 solubility with increasing dilution. Purified tankyrase was diluted to 16.7, 5.6, 1.9, 0.62, or 0.2 μg/ml prior to centrifugation, and protein in the supernatant (S) or pellet (P) was detected by Western blotting with tankyrase antibody. (C) TRF1 binding to tankyrase polymers. His-tagged TRF1 was incubated with or without purified His-tagged tankyrase1 (TNK) or tankyrase1-ΔSAM (ΔSAM) and then centrifuged. Soluble and pelleted fractions were analyzed by Western blotting with His antibody. The arrow points to tankyrase 1 or tankyrase1-ΔSAM; the arrowhead indicates TRF1. (D) PARP activity of recombinant tankyrase 1. Tankyrase (upper bracket) or TRF1 (lower bracket) poly(ADP-ribosyl)ation was detected by Western blotting with antibody to poly(ADP-ribose). The arrow marks the input TRF1, which started with a low level of modification. This was probably caused by a copurifying PARP activity. The proteins present in each reaction mixture are indicated at the top; the presence (+) or absence (−) of 1 mM NAD and 10 mM 3-aminobenzamide (3AB) is indicated.
FIG. 3.
In vivo complex formation. (A) Solubility of overexpressed tankyrase 1 in LMH cell lysates. Cells expressing FL-tankyrase1 (TNK) or tankyrase1-ΔSAM (ΔSAM) were lysed at 4 or 25°C and centrifuged, and tankyrase was detected in the supernatant (S) or pellet (P) by Western blotting with tankyrase antibody. (B) Superose 6 fractionation of soluble tankyrase extracted from cells expressing FL-tankyrase1 (TNK) or tankyrase1-ΔSAM (ΔSAM) or from untransfected cells (endo). The total amount of protein loaded on the column was ∼250 μg from TNK- and ΔSAM-expressing cells and ∼650 μg from wild-type cells. The bands corresponding to the input material are shown in the lane to the left; the amounts loaded were 1% of the total for TNK, 2% for SAM, and 10% for wild-type extracts. The peak fractions containing marker proteins are shown at the top, and fraction numbers are shown below. Detection was by Western blotting with tankyrase antibody.
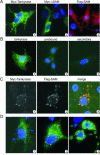
FIG. 4.
In vivo tankyrase localization. (A) LMH cells expressing Myc-tagged FL-tankyrase1 (Myc-Tankyrase), Myc-tagged tankyrase1-ΔSAM (Myc-ΔSAM), or Flag-tagged SAM domain (Flag-SAM) were fixed and incubated with antibody to the Myc or Flag tags. Nuclei were counterstained with DAPI. (B) Untransfected cells were stained with antibody to the tankyrase SAM domain (panel 1), tankyrase antibody that had been preincubated with purified SAM domain (panel 2), or secondary antibody (panel 3). (C) Cells coexpressing Myc-tagged FL-tankyrase1 and Flag-tagged SAM domain. Panel 1, staining with Myc antibody; panel 2, staining with Flag antibody; panel 3, combination of the Myc (green) and Flag (red) signals. (D) Examples of the different size of vesicle-like structures observed in FL-tankyrase1-expressing cells. Detection was with the Myc antibody.
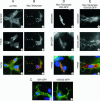
FIG. 5.
Effects of tankyrase overexpression on the secretory pathway. (A and B) Colocalization of tankyrase vesicles with FTCD. Control LMH cells (A) or LMH cells expressing Myc-tagged FL-tankyrase1 (B) were fixed and incubated with tankyrase antibody (A1), Myc antibody (B1), or FTCD antibody (A2 and B2). Panels A3 and B3 show an overlay of the tankyrase (green) and FTCD (red) signals. Cells were counterstained with DAPI. wt, wild type. (C and D) Effect of Myc-FL-tankyrase1 on secretion of marker proteins. LMH cells were transfected with GPI-GFP or VSVG3-GFP and analyzed by fluorescence microscopy after 30 h at 37°C (GPI-GFP) or 32°C (the permissive temperature for temperature-sensitive VSVG3-GFP). (C) Localization of GPI-GFP and VSVG3-GFP in wild-type cells. (D and E) Localization of GPI-GFP and VSVG3-GFP after tankyrase overexpression. Cells were transfected with Myc-tagged FL-tankyrase1 and GPI-GFP or VSVG3-GFP. (D1 and E1) Exogenous tankyrase detected with Myc antibody; (D2 and E2) GPI-GFP and VSVG3-GFP staining; (D3 and E3) overlay of panels 1 and 2. Red, tankyrase; green, GPI-GFP and VSVG3-GFP.
FIG. 6.
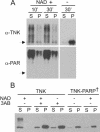
Dissociation of tankyrase complexes by tankyrase autoribosylation. (A) Solubility of purified full-length tankyrase 1 after incubation with NAD+. Samples were incubated with (+) or without (−) 1 mM NAD+ for 10 or 30 min at 25°C and centrifuged to separate soluble (S) from insoluble (P) material. Tankyrase was detected by Western blotting with antibody to tankyrase (α-TNK) or poly(ADP-ribose) (α-PAR). Arrow, position of unmodified tankyrase. (B) Solubility of overexpressed tankyrase 1 in LMH cell extracts. Cells expressing FL-tankyrase1 (TNK) or catalytically inactive FL-tankyrase1 (TNK-PARP†) were lysed at 25°C in the presence (+) or absence (−) of 1 mM NAD+ and/or 10 mM 3AB and centrifuged. Tankyrase was detected in the supernatant (S) or pellet (P) by Western blotting with tankyrase antibody.
FIG. 7.
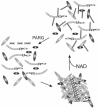
Model for tankyrase lattice assembly and disassembly. Lattice assembly occurs through SAM domain-mediated self-association of tankyrase molecules and binding of tankyrase interaction partners to the individual tankyrase ankyrin repeat modules. Tankyrase autoribosylation and poly(ADP-ribosyl)ation of interaction partners causes lattice disassembly. Subsequent removal of the poly(ADP-ribose) chains by PARG would then allow lattice reassembly.
Similar articles
- Vertebrate tankyrase domain structure and sterile alpha motif (SAM)-mediated multimerization.
De Rycker M, Venkatesan RN, Wei C, Price CM. De Rycker M, et al. Biochem J. 2003 May 15;372(Pt 1):87-96. doi: 10.1042/BJ20021450. Biochem J. 2003. PMID: 12589701 Free PMC article. - Tankyrase Sterile α Motif Domain Polymerization Is Required for Its Role in Wnt Signaling.
Riccio AA, McCauley M, Langelier MF, Pascal JM. Riccio AA, et al. Structure. 2016 Sep 6;24(9):1573-81. doi: 10.1016/j.str.2016.06.022. Epub 2016 Aug 4. Structure. 2016. PMID: 27499439 Free PMC article. - Identification of a novel human tankyrase through its interaction with the adaptor protein Grb14.
Lyons RJ, Deane R, Lynch DK, Ye ZS, Sanderson GM, Eyre HJ, Sutherland GR, Daly RJ. Lyons RJ, et al. J Biol Chem. 2001 May 18;276(20):17172-80. doi: 10.1074/jbc.M009756200. Epub 2001 Feb 22. J Biol Chem. 2001. PMID: 11278563 - Tankyrases as drug targets.
Lehtiö L, Chi NW, Krauss S. Lehtiö L, et al. FEBS J. 2013 Aug;280(15):3576-93. doi: 10.1111/febs.12320. Epub 2013 Jun 18. FEBS J. 2013. PMID: 23648170 Review. - Poly(ADP-ribose) polymerase enzymes and the maintenance of genome integrity.
Eisemann T, Pascal JM. Eisemann T, et al. Cell Mol Life Sci. 2020 Jan;77(1):19-33. doi: 10.1007/s00018-019-03366-0. Epub 2019 Nov 21. Cell Mol Life Sci. 2020. PMID: 31754726 Free PMC article. Review.
Cited by
- The zinc-binding motif in tankyrases is required for the structural integrity of the catalytic ADP-ribosyltransferase domain.
Sowa ST, Lehtiö L. Sowa ST, et al. Open Biol. 2022 Mar;12(3):210365. doi: 10.1098/rsob.210365. Epub 2022 Mar 23. Open Biol. 2022. PMID: 35317661 Free PMC article. - Multiple E3 ligases control tankyrase stability and function.
Perrard J, Smith S. Perrard J, et al. Nat Commun. 2023 Nov 8;14(1):7208. doi: 10.1038/s41467-023-42939-3. Nat Commun. 2023. PMID: 37938264 Free PMC article. - The PARsylation activity of tankyrase in adipose tissue modulates systemic glucose metabolism in mice.
Zhong L, Ding Y, Bandyopadhyay G, Waaler J, Börgeson E, Smith S, Zhang M, Phillips SA, Mahooti S, Mahata SK, Shao J, Krauss S, Chi NW. Zhong L, et al. Diabetologia. 2016 Mar;59(3):582-91. doi: 10.1007/s00125-015-3815-1. Epub 2015 Dec 2. Diabetologia. 2016. PMID: 26631215 - Cross-species difference in telomeric function of tankyrase 1.
Muramatsu Y, Ohishi T, Sakamoto M, Tsuruo T, Seimiya H. Muramatsu Y, et al. Cancer Sci. 2007 Jun;98(6):850-7. doi: 10.1111/j.1349-7006.2007.00462.x. Epub 2007 Apr 13. Cancer Sci. 2007. PMID: 17433040 Free PMC article. - The DNA damage-inducible C. elegans tankyrase is a nuclear protein closely linked to chromosomes.
White C, Gagnon SN, St-Laurent JF, Gravel C, Proulx LI, Desnoyers S. White C, et al. Mol Cell Biochem. 2009 Apr;324(1-2):73-83. doi: 10.1007/s11010-008-9986-z. Epub 2008 Dec 23. Mol Cell Biochem. 2009. PMID: 19104912
References
- Bae, J., J. R. Donigian, and A. J. Hsueh. 2003. Tankyrase 1 interacts with Mcl-1 proteins and inhibits their regulation of apoptosis. J. Biol. Chem. 278:5195-5204. - PubMed
- Bennett, V., and A. J. Baines. 2001. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol. Rev. 81:1353-1392. - PubMed
- Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. - PubMed
- Boeckers, T. M., J. Bockmann, M. R. Kreutz, and E. D. Gundelfinger. 2002. ProSAP/Shank proteins-a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J. Neurochem. 81:903-910. - PubMed
- Bryant, N. J., R. Govers, and D. E. James. 2002. Regulated transport of the glucose transporter GLUT4. Nat. Rev. Mol. Cell Biol. 3:267-277. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources