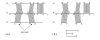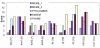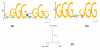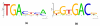PhyME: a probabilistic algorithm for finding motifs in sets of orthologous sequences - PubMed (original) (raw)
Comparative Study
PhyME: a probabilistic algorithm for finding motifs in sets of orthologous sequences
Saurabh Sinha et al. BMC Bioinformatics. 2004.
Abstract
Background: This paper addresses the problem of discovering transcription factor binding sites in heterogeneous sequence data, which includes regulatory sequences of one or more genes, as well as their orthologs in other species.
Results: We propose an algorithm that integrates two important aspects of a motif's significance - overrepresentation and cross-species conservation - into one probabilistic score. The algorithm allows the input orthologous sequences to be related by any user-specified phylogenetic tree. It is based on the Expectation-Maximization technique, and scales well with the number of species and the length of input sequences. We evaluate the algorithm on synthetic data, and also present results for data sets from yeast, fly, and human.
Conclusions: The results demonstrate that the new approach improves motif discovery by exploiting multiple species information.
Figures
Figure 1
Orthologous promoters and blocks of sequence conservation. Shaded areas represent ungapped aligned blocks. _σ_1 is the reference species. (a) Alignment of input sequences and extraction of blocks. (b) Reorganization of input sequences.
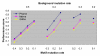
Figure 2
Effect of varying the number of species (K) on motif-finding performance. The x-axis is the relative entropy (R) of the planted motif. Each point is an average over 10 experiments with synthetic data. (μ b = 0.3, μ m = 0.1.)
Figure 3
Effect of varying background and motif mutation rates (μ b and μ m respectively) on motif-finding performance. Each point is an average over 10 experiments with synthetic data. (K = 3, R = 12.)
Figure 4
Effect of the alignment step on motif-finding performance. The x-axis shows how many of the orthologous pairs of planted motifs are artificially unpaired in the alignment step. Each solid line represents a separate experiment. The squares plot the average score over eight experiments.
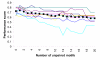
Figure 5
Effect of multiple species information on motif-discovery in the regulons RAP1, MIG1, CAR1, PHO4 and MCM1 in yeast. The y-axis plots the number of matches to the known motif, among the top η reported occurrences, where η is the number of known sites, plotted as "KNOWN" . Only matches in S. cerevisiae are considered.
Figure 6
Comparison of PhyME to 1 species and 2 species MEME, and to PhyloGibbs and EMnEM, for fly enhancers. The parenthetical number next to an enhancer name is the number of strong occurrences of the known weight matrix, in the D. melanogaster sequence.
Figure 7
Results on the human SP1 regulon. (a) The known motif. (b) Motif reported by PhyME, using mouse and rat orthologs. (c) The phylogenetic tree used by PhyME.
Figure 8
Results on the human c-Jun regulon. (a) The known motif. (b) Motif reported by PhyME, using mouse and rat orthologs.
Similar articles
- PhyloGibbs: a Gibbs sampling motif finder that incorporates phylogeny.
Siddharthan R, Siggia ED, van Nimwegen E. Siddharthan R, et al. PLoS Comput Biol. 2005 Dec;1(7):e67. doi: 10.1371/journal.pcbi.0010067. Epub 2005 Dec 9. PLoS Comput Biol. 2005. PMID: 16477324 Free PMC article. - Phylogenetic motif detection by expectation-maximization on evolutionary mixtures.
Moses AM, Chiang DY, Eisen MB. Moses AM, et al. Pac Symp Biocomput. 2004:324-35. doi: 10.1142/9789812704856_0031. Pac Symp Biocomput. 2004. PMID: 14992514 - PhyME: a software tool for finding motifs in sets of orthologous sequences.
Sinha S. Sinha S. Methods Mol Biol. 2007;395:309-18. Methods Mol Biol. 2007. PMID: 17993682 - Detecting DNA regulatory motifs by incorporating positional trends in information content.
Kechris KJ, van Zwet E, Bickel PJ, Eisen MB. Kechris KJ, et al. Genome Biol. 2004;5(7):R50. doi: 10.1186/gb-2004-5-7-r50. Epub 2004 Jun 24. Genome Biol. 2004. PMID: 15239835 Free PMC article. - Finding regulatory elements and regulatory motifs: a general probabilistic framework.
van Nimwegen E. van Nimwegen E. BMC Bioinformatics. 2007 Sep 27;8 Suppl 6(Suppl 6):S4. doi: 10.1186/1471-2105-8-S6-S4. BMC Bioinformatics. 2007. PMID: 17903285 Free PMC article. Review.
Cited by
- A Monte Carlo-based framework enhances the discovery and interpretation of regulatory sequence motifs.
Seitzer P, Wilbanks EG, Larsen DJ, Facciotti MT. Seitzer P, et al. BMC Bioinformatics. 2012 Nov 27;13:317. doi: 10.1186/1471-2105-13-317. BMC Bioinformatics. 2012. PMID: 23181585 Free PMC article. - Large-scale cis-element detection by analysis of correlated expression and sequence conservation between Arabidopsis and Brassica oleracea.
Haberer G, Mader MT, Kosarev P, Spannagl M, Yang L, Mayer KF. Haberer G, et al. Plant Physiol. 2006 Dec;142(4):1589-602. doi: 10.1104/pp.106.085639. Epub 2006 Oct 6. Plant Physiol. 2006. PMID: 17028152 Free PMC article. - iCR: a web tool to identify conserved targets of a regulatory protein across the multiple related prokaryotic species.
Ranjan S, Seshadri J, Vindal V, Yellaboina S, Ranjan A. Ranjan S, et al. Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W584-7. doi: 10.1093/nar/gkl202. Nucleic Acids Res. 2006. PMID: 16845075 Free PMC article. - Limitations and potentials of current motif discovery algorithms.
Hu J, Li B, Kihara D. Hu J, et al. Nucleic Acids Res. 2005 Sep 2;33(15):4899-913. doi: 10.1093/nar/gki791. Print 2005. Nucleic Acids Res. 2005. PMID: 16284194 Free PMC article. - Reliable prediction of transcription factor binding sites by phylogenetic verification.
Li X, Zhong S, Wong WH. Li X, et al. Proc Natl Acad Sci U S A. 2005 Nov 22;102(47):16945-50. doi: 10.1073/pnas.0504201102. Epub 2005 Nov 14. Proc Natl Acad Sci U S A. 2005. PMID: 16286651 Free PMC article.
References
- Bailey TL, Elkan C. Unsupervised learning of multiple motifs in biopolymers using expectation maximization. Machine Learning. 1995;21:51–80. doi: 10.1023/A:1022617714621. - DOI
- Hertz GZ, Hartzell GW, III, Stormo GD. Identification of Consensus Patterns in Unaligned DNA Sequences Known to be Functionally Related. Computer Applications in the Biosciences. 1990;6:81–92. - PubMed
- Lawrence CE, Altschul SF, Boguski MS, Liu JS, Neuwald AF, Wootton JC. Detecting Subtle Sequence Signals: a Gibbs Sampling Strategy for Multiple Alignment. Science. 1993;262:208–214. - PubMed
- Sinha S, Tompa M. A Statistical Method for Finding Transcription Factor Binding Sites. In Proceedings of the Eigth International Conference on Intelligent Systems for Molecular Biology: August 2000; La Jolla. 2000. pp. 344–354. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases