Natural genetic variation caused by transposable elements in humans - PubMed (original) (raw)
Natural genetic variation caused by transposable elements in humans
E Andrew Bennett et al. Genetics. 2004 Oct.
Abstract
Transposons and transposon-like repetitive elements collectively occupy 44% of the human genome sequence. In an effort to measure the levels of genetic variation that are caused by human transposons, we have developed a new method to broadly detect transposon insertion polymorphisms of all kinds in humans. We began by identifying 606,093 insertion and deletion (indel) polymorphisms in the genomes of diverse humans. We then screened these polymorphisms to detect indels that were caused by de novo transposon insertions. Our method was highly efficient and led to the identification of 605 nonredundant transposon insertion polymorphisms in 36 diverse humans. We estimate that this represents 25-35% of approximately 2075 common transposon polymorphisms in human populations. Because we identified all transposon insertion polymorphisms with a single method, we could evaluate the relative levels of variation that were caused by each transposon class. The average human in our study was estimated to harbor 1283 Alu insertion polymorphisms, 180 L1 polymorphisms, 56 SVA polymorphisms, and 17 polymorphisms related to other forms of mobilized DNA. Overall, our study provides significant steps toward (i) measuring the genetic variation that is caused by transposon insertions in humans and (ii) identifying the transposon copies that produce this variation.
Figures
Figure 1.—
Computational pipeline for indel and transposon polymorphism discovery. A flow chart of the computational steps that were followed for the discovery of indel and transposon polymorphisms is shown on the left (boxes). A breakdown of the number of traces present at the end of each step is shown on the right. The number of indel and transposon polymorphisms identified is listed at the bottom. Note that the numbers are broken down for each of the three populations examined. TSC, the SNP Consortium traces; WGS, whole-genome shotgun traces; WCS, whole-chromosome shotgun traces.
Figure 2.—
Proposed new evolutionary lineages of Alu. For each Alu subfamily, the number of polymorphic copies retaining the subfamily consensus sequence is compared to groups sharing one or more base pair changes (in parentheses). In several cases, the majority of polymorphic copies of a given subfamily diverge from the subfamily consensus by a few shared changes. An evolutionary progression can be inferred (from left to right) in which new shared base changes appear to have been acquired. The 10 proposed novel evolutionary groups are indicated by braces to the right of the groups. The total number of elements within the group is indicated in the parentheses. These data suggest that a significant number of Alu insertions in the genome can serve as new source genes to produce offspring elements. Only elements that are at least 80% full length are represented.
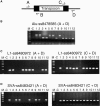
Figure 3.—
PCR validation studies. The strategy for the PCR validation assays is shown along with some examples of these assays. (A) The locations of the four primers (A, B, C, and D) that were used to evaluate transposon polymorphisms by PCR are shown. (B) A typical Alu PCR assay is shown in which primers flanking the transposon (A and D) are used to determine whether a given Alu copy is present in the genome of an individual. The larger band is produced when the element is present, whereas the smaller band is produced when the element is absent. Lane M, 1-kb marker; −C, negative control lacking template DNA; lanes 1–12, 12 PCR reactions evaluating DNA samples from the Coriell panel. (C) A typical L1 PCR assay is shown in which two PCRs are performed to identify all of the alleles present in the Coriell panel. The assay on the left uses the A and D primers to identify alleles that lack an L1 insertion, whereas the assay on the right uses primers C and D (or A and B) to identify alleles containing the L1 insertion. These two assays are used together to evaluate whether a given allele is homozygous or heterozygous in a given individual. The lanes are the same as for B. For cases in which the L1 element is relatively short (<2 kb), the allele containing the L1 insertion often was detected in the A plus D assay as well. (D) A typical SVA assay is depicted. These assays are performed the same way as the L1 assays (in C above), using two assays to evaluate all SVA alleles present.
Similar articles
- Recently mobilized transposons in the human and chimpanzee genomes.
Mills RE, Bennett EA, Iskow RC, Luttig CT, Tsui C, Pittard WS, Devine SE. Mills RE, et al. Am J Hum Genet. 2006 Apr;78(4):671-9. doi: 10.1086/501028. Epub 2006 Feb 2. Am J Hum Genet. 2006. PMID: 16532396 Free PMC article. - Mobile elements create structural variation: analysis of a complete human genome.
Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, Zhou Q, Kirkness EF, Levy S, Batzer MA, Jorde LB. Xing J, et al. Genome Res. 2009 Sep;19(9):1516-26. doi: 10.1101/gr.091827.109. Epub 2009 May 13. Genome Res. 2009. PMID: 19439515 Free PMC article. - Which transposable elements are active in the human genome?
Mills RE, Bennett EA, Iskow RC, Devine SE. Mills RE, et al. Trends Genet. 2007 Apr;23(4):183-91. doi: 10.1016/j.tig.2007.02.006. Epub 2007 Feb 27. Trends Genet. 2007. PMID: 17331616 Review. - Human transposon tectonics.
Burns KH, Boeke JD. Burns KH, et al. Cell. 2012 May 11;149(4):740-52. doi: 10.1016/j.cell.2012.04.019. Cell. 2012. PMID: 22579280 Free PMC article. Review.
Cited by
- Comparative Genomics of Fungi in Nectriaceae Reveals Their Environmental Adaptation and Conservation Strategies.
Rissi DV, Ijaz M, Baschien C. Rissi DV, et al. J Fungi (Basel). 2024 Sep 5;10(9):632. doi: 10.3390/jof10090632. J Fungi (Basel). 2024. PMID: 39330392 Free PMC article. - Transposable element insertions have strongly affected human evolution.
Britten RJ. Britten RJ. Proc Natl Acad Sci U S A. 2010 Nov 16;107(46):19945-8. doi: 10.1073/pnas.1014330107. Epub 2010 Nov 1. Proc Natl Acad Sci U S A. 2010. PMID: 21041622 Free PMC article. - The minimal active human SVA retrotransposon requires only the 5'-hexamer and Alu-like domains.
Hancks DC, Mandal PK, Cheung LE, Kazazian HH Jr. Hancks DC, et al. Mol Cell Biol. 2012 Nov;32(22):4718-26. doi: 10.1128/MCB.00860-12. Epub 2012 Sep 24. Mol Cell Biol. 2012. PMID: 23007156 Free PMC article. - A streamlined method for detecting structural variants in cancer genomes by short read paired-end sequencing.
Mijušković M, Brown SM, Tang Z, Lindsay CR, Efstathiadis E, Deriano L, Roth DB. Mijušković M, et al. PLoS One. 2012;7(10):e48314. doi: 10.1371/journal.pone.0048314. Epub 2012 Oct 29. PLoS One. 2012. PMID: 23144753 Free PMC article.
References
- Abdel-Halim, S., G. E. Kilroy, W. S. Watkins, L. B. Jorde and M. A. Batzer, 2003. Recently integrated Alu elements and human genomic diversity. Mol. Biol. Evol. 20: 1349–1361. - PubMed
- Bailey, J. A., Z. Gu, R. A. Clark, K. Reinert, R. V. Samonte et al., 2002. Recent segmental duplications in the human genome. Science 297: 1003–1007. - PubMed
- Batzer, M. A., and P. L. Deininger, 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3: 370–379. - PubMed
- Bedell, J. A., I. Korf and W. Gish, 2000. MaskerAid: a performance enhancement to RepeatMasker. Bioinformatics 16: 1040–1041. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HG002898/HG/NHGRI NIH HHS/United States
- T32 GM008490/GM/NIGMS NIH HHS/United States
- 2T32GM008490-11/GM/NIGMS NIH HHS/United States
- 1R01HG02898-01A1/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials