Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure - PubMed (original) (raw)
Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure
Winfried Denk et al. PLoS Biol. 2004 Nov.
Abstract
Three-dimensional (3D) structural information on many length scales is of central importance in biological research. Excellent methods exist to obtain structures of molecules at atomic, organelles at electron microscopic, and tissue at light-microscopic resolution. A gap exists, however, when 3D tissue structure needs to be reconstructed over hundreds of micrometers with a resolution sufficient to follow the thinnest cellular processes and to identify small organelles such as synaptic vesicles. Such 3D data are, however, essential to understand cellular networks that, particularly in the nervous system, need to be completely reconstructed throughout a substantial spatial volume. Here we demonstrate that datasets meeting these requirements can be obtained by automated block-face imaging combined with serial sectioning inside the chamber of a scanning electron microscope. Backscattering contrast is used to visualize the heavy-metal staining of tissue prepared using techniques that are routine for transmission electron microscopy. Low-vacuum (20-60 Pa H(2)O) conditions prevent charging of the uncoated block face. The resolution is sufficient to trace even the thinnest axons and to identify synapses. Stacks of several hundred sections, 50-70 nm thick, have been obtained at a lateral position jitter of typically under 10 nm. This opens the possibility of automatically obtaining the electron-microscope-level 3D datasets needed to completely reconstruct the connectivity of neuronal circuits.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures
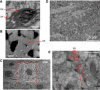
Figure 1. Resolution and Contrast Using the Backscattered Electron Signal
(A and B) Presynaptic vesicles (SV) and postsynaptic folds (SF) are clearly visible (A) in a motor endplate preparation embedded in Spurr's resin. Similarly, the hexagonal array of actin filaments (AA) can be clearly resolved (B) in a different region from the same image (both images were smoothed using the ImageJ “smooth” command). Imaging conditions for (A) and (B): electron energy, 7.5 keV; spot, 3.5; chamber pressure, 30 Pa (H2O); pixel dwell time, 30 μs. The scanning resolution was 6.7 nm/pixel. (C) The effect of beam exposure on the block surface. Note the increased brightness and the lack of chatter in the central region (inside the dashed rectangle), from which a stack was acquired at higher resolution before taking the image shown. The tissue was rat neocortex embedded in Spurr's resin. Imaging conditions for (C): electron energy, 7.5 keV; spot, 3; digital resolution for stack acquisition, 26.7 nm/pixel; dwell time, 30 μs. (D and E) Cortical tissue embedded in Epon. Synapses (SD) are clearly discernable (E). Imaging conditions for (D) and (E): electron energy, 7.5 keV beam current; spot, 3; chamber pressure, 30Pa (H2O); pixel dwell time, 30 μs. The scanning resolution was 9.5 nm/pixel. Note that more backscattering corresponds to darker pixels in (A), (B), (D), and (E) but to brighter pixels in (C).
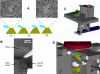
Figure 2. SEM Microtomy
(A) Principle of SBFSEM operation: (1) a SEM image is taken of the surface of the plastic-embedded tissue preparation (amber trapezoid). (2) Then with a diamond knife (blue) an ultrathin slice is cut off the top of the block. (3) After retraction of the knife, the next picture is taken. The pictures shown are from an actual stack (cerebellar cortex) but are not successive slices; rather, they are spaced by five images (about 315 nm) to make the changes more apparent. (B) Usually cut-off slices pile up on the top of the knife. Protruding into the picture from the right is a puffer pipette, occasionally used to remove debris from the knife. (C and D) The mechanical design for the in-chamber microtome is shown in an overview (C) and a close-up of knife and sample (D) in renderings from the computer-aided design software. Most parts are nonmagnetic stainless steel (grey). A large-motion leveraged piezo actuator (green part on the left) drives the knife holder back and forth. The custom diamond knife (light blue) is clamped in a special holder. The sample (amber) advance is driven via a lever by a direct-current-motor-driven micrometer (dark blue). The retraction during the backwards knife motion is again piezo actuated (green cylinder in the lower right of [C]). Bearing springs are brown. The BSE detector (red) is depicted schematically above the sample. Not shown is the lateral positioning mechanism.
Figure 3. 3D Datasets: Five Slices
(A) Five slices from a stack containing a total of 365 slices at 63-nm section thickness; same tissue as in Figure 1A and 1B. Note beginning of myelin sheath (MS) in the lowest slice. Imaging conditions as in Figures 1A and 1B except resolution is 13.4 nm/pixel. (B) Reslice of the same dataset along the red line show in the top image of (A). Arrows point to the presynaptic ending (PS) and the z-band (ZB). Image corners in (A) touch the reslice (B) at the depths at which they were taken. (C and D) Cerebellar tissue displayed at low (C) and high (D) resolution; note nuclear envelope (NM). Note differences in lateral and vertical resolution. Imaging conditions for (C) and (D): electron energy, 7.5 keV; spot size, 3.5; digital resolution, 12.7 nm/pixel.
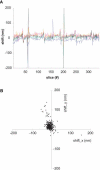
Figure 4. The Alignment of Successive Images in a Stack
Shifts between images were quantified using the positions of the peaks of the cross correlation (see Materials and Methods); same dataset as in Figure 3A and 3B. (A) The peak shifts in x-direction are shown for five different subregions distributed over the field of view. Four of the regions have a size of 256 × 256 pixels, one has a size of 512 × 512 (black trace). The peaks around slices 59 and 202 are caused by slice debris on the block face (see also streaks in Figure 3B). (B) The x/y displacement for the 512 × 512 region is shown in a scatter plot. For the central cluster the standard deviations are 10.9 nm and 11.8 nm for x and y, respectively.
Figure 5. Energy Dependence of the Depth Resolution
The lateral resolution does not change very much as the electron energy is reduced from 7.5 keV (A) to 4 keV (B), but the resolution along the z-direction is dramatically different (C). The lines between (A), (B), and (C) indicate the z-positions in the stack from which (A) and (B) were taken. In the high-resolution view (lower panel in [C]) membranes that were en-face (EM) in the original slices can be clearly recognized. The nominal slice thickness was 55 nm. Tissue is rat cortex. Imaging conditions: dwell time, 25 μs; spot sizes, 3.5 and 2.8 for 4 and 7.5 keV, respectively.
Similar articles
- Developing 3D SEM in a broad biological context.
Kremer A, Lippens S, Bartunkova S, Asselbergh B, Blanpain C, Fendrych M, Goossens A, Holt M, Janssens S, Krols M, Larsimont JC, Mc Guire C, Nowack MK, Saelens X, Schertel A, Schepens B, Slezak M, Timmerman V, Theunis C, VAN Brempt R, Visser Y, Guérin CJ. Kremer A, et al. J Microsc. 2015 Aug;259(2):80-96. doi: 10.1111/jmi.12211. Epub 2015 Jan 26. J Microsc. 2015. PMID: 25623622 Free PMC article. - Knife-edge scanning microscopy for imaging and reconstruction of three-dimensional anatomical structures of the mouse brain.
Mayerich D, Abbott L, McCormick B. Mayerich D, et al. J Microsc. 2008 Jul;231(Pt 1):134-43. doi: 10.1111/j.1365-2818.2008.02024.x. J Microsc. 2008. PMID: 18638197 - Combining serial block face and focused ion beam scanning electron microscopy for 3D studies of rare events.
Guérin CJ, Kremer A, Borghgraef P, Shih AY, Lippens S. Guérin CJ, et al. Methods Cell Biol. 2019;152:87-101. doi: 10.1016/bs.mcb.2019.03.014. Epub 2019 May 7. Methods Cell Biol. 2019. PMID: 31326028 - New developments in electron microscopy for serial image acquisition of neuronal profiles.
Kubota Y. Kubota Y. Microscopy (Oxf). 2015 Feb;64(1):27-36. doi: 10.1093/jmicro/dfu111. Epub 2015 Jan 5. Microscopy (Oxf). 2015. PMID: 25564566 Review. - Challenges of microtome-based serial block-face scanning electron microscopy in neuroscience.
Wanner AA, Kirschmann MA, Genoud C. Wanner AA, et al. J Microsc. 2015 Aug;259(2):137-142. doi: 10.1111/jmi.12244. Epub 2015 Apr 23. J Microsc. 2015. PMID: 25907464 Free PMC article. Review.
Cited by
- Three-dimensional ultrastructure of the brain pericyte-endothelial interface.
Ornelas S, Berthiaume AA, Bonney SK, Coelho-Santos V, Underly RG, Kremer A, Guérin CJ, Lippens S, Shih AY. Ornelas S, et al. J Cereb Blood Flow Metab. 2021 Sep;41(9):2185-2200. doi: 10.1177/0271678X211012836. Epub 2021 May 10. J Cereb Blood Flow Metab. 2021. PMID: 33970018 Free PMC article. - Mitochondrial reticulum for cellular energy distribution in muscle.
Glancy B, Hartnell LM, Malide D, Yu ZX, Combs CA, Connelly PS, Subramaniam S, Balaban RS. Glancy B, et al. Nature. 2015 Jul 30;523(7562):617-20. doi: 10.1038/nature14614. Nature. 2015. PMID: 26223627 Free PMC article. - Volume electron microscopy: analyzing the lung.
Schneider JP, Hegermann J, Wrede C. Schneider JP, et al. Histochem Cell Biol. 2021 Feb;155(2):241-260. doi: 10.1007/s00418-020-01916-3. Epub 2020 Sep 17. Histochem Cell Biol. 2021. PMID: 32944795 Free PMC article. Review. - Illuminating the Brain With X-Rays: Contributions and Future Perspectives of High-Resolution Microtomography to Neuroscience.
Rodrigues PV, Tostes K, Bosque BP, de Godoy JVP, Amorim Neto DP, Dias CSB, Fonseca MC. Rodrigues PV, et al. Front Neurosci. 2021 Mar 17;15:627994. doi: 10.3389/fnins.2021.627994. eCollection 2021. Front Neurosci. 2021. PMID: 33815039 Free PMC article. Review. - Three-dimensional structure analysis and percolation properties of a barrier marine coating.
Chen B, Guizar-Sicairos M, Xiong G, Shemilt L, Diaz A, Nutter J, Burdet N, Huo S, Mancuso J, Monteith A, Vergeer F, Burgess A, Robinson I. Chen B, et al. Sci Rep. 2013;3:1177. doi: 10.1038/srep01177. Epub 2013 Jan 31. Sci Rep. 2013. PMID: 23378910 Free PMC article.
References
- Alkemper J, Voorhees PW. Quantitative serial sectioning analysis. J Microsc. 2001;201:388–394. - PubMed
- Ardenne M. Das Elektronen-Rastermikroskop. Theoretische Grundlagen. Z Phys. 1938a;109:553–572.
- Ardenne M. Das Elektronen-Rastermikroskop. Praktische Ausfuehrung. Z Tech Phys. 1938b;19:407–416.
- Baumeister W. Electron tomography: Towards visualizing the molecular organization of the cytoplasm. Curr Opin Struct Biol. 2002;12:679–684. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous