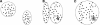Whole-genome analysis of Alu repeat elements reveals complex evolutionary history - PubMed (original) (raw)
Whole-genome analysis of Alu repeat elements reveals complex evolutionary history
Alkes L Price et al. Genome Res. 2004 Nov.
Abstract
Alu repeats are the most abundant family of repeats in the human genome, with over 1 million copies comprising 10% of the genome. They have been implicated in human genetic disease and in the enrichment of gene-rich segmental duplications in the human genome, and they form a rich fossil record of primate and human history. Alu repeat elements are believed to have arisen from the replication of a small number of source elements, whose evolution over time gives rise to the 31 Alu subfamilies currently reported in Repbase Update. We apply a novel method to identify and statistically validate 213 Alu subfamilies. We build an evolutionary tree of these subfamilies and conclude that the history of Alu evolution is more complex than previous studies had indicated.
Figures
Figure 1.
Applicability of _k_-means clustering to different kinds of clustering problems. Disjoint clusters of similar size are easily identified (A). Small subfamilies nested inside large subfamilies, a typical scenario in Alu repeat subfamilies, are not easily identified, because there is a tendency to split off a larger cluster (B) instead of identifying the nested subfamily (C).
Figure 2.
Aligned consensus sequences of selected subfamilies. (Top) The consensus sequence of the entire Alu family, with positions labeled from 1 to 282. (Middle) The consensus sequences of six Alu subfamilies we identified that are currently reported in Repbase Update: _Alu_Jo, _Alu_Sx, _Alu_Sq, _Alu_Sp, _Alu_Y, and _Alu_Ya5; the few discrepancies between our consensus sequences and the consensus sequences reported in Repbase Update occur mostly at CpG dinucleotide positions, which are ill-determined because of frequent mutation. (Bottom) The consensus sequences of six Alu subfamilies we identified that are not currently reported in Repbase Update: _Alu_Sx_3, _Alu_Sx_5, _Alu_Sq_3, _Alu_Sg_4, _Alu_Sc_8, and _Alu_Y_8.
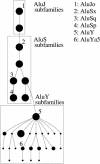
Figure 3.
Evolutionary tree of the 31 subfamilies currently reported in Repbase Update. (Large nodes) Subfamilies with more than 10,000 elements; (medium nodes) 1000 to 10,000 elements; (small nodes) less than 1000 elements. Each of the 6 Repbase Update subfamilies listed in Figure 2 is labeled. The _Alu_J, _Alu_S, and _Alu_Y classes of subfamilies are contained in boxes.
Figure 4.
Evolutionary tree of the 213 subfamilies we identified. (Large nodes) Subfamilies with more than 10,000 elements; (medium nodes) 1000 to 10,000 elements; (small nodes) less than 1000 elements. Subfamilies listed in Repbase Update are colored blue, and the 6 novel subfamilies listed in Figure 2 are colored red. Each of the subfamilies listed in Figure 2 is labeled. A rendition of this tree with every node labeled is available in the Supplementary materials online. The _Alu_J, _Alu_S, and _Alu_Y classes of subfamilies are contained in boxes; not all subfamilies fit into one of these classes. A timeline roughly depicting the average divergence of each subfamily from its consensus sequence and the approximate age obtained by applying a constant scaling factor of 4 million years per 1% divergence from consensus sequence are included at right.
Similar articles
- An Alu transposition model for the origin and expansion of human segmental duplications.
Bailey JA, Liu G, Eichler EE. Bailey JA, et al. Am J Hum Genet. 2003 Oct;73(4):823-34. doi: 10.1086/378594. Epub 2003 Sep 22. Am J Hum Genet. 2003. PMID: 14505274 Free PMC article. - Impact of Alu repeats on the evolution of human p53 binding sites.
Cui F, Sirotin MV, Zhurkin VB. Cui F, et al. Biol Direct. 2011 Jan 6;6:2. doi: 10.1186/1745-6150-6-2. Biol Direct. 2011. PMID: 21208455 Free PMC article. - Recently integrated Alu elements and human genomic diversity.
Salem AH, Kilroy GE, Watkins WS, Jorde LB, Batzer MA. Salem AH, et al. Mol Biol Evol. 2003 Aug;20(8):1349-61. doi: 10.1093/molbev/msg150. Epub 2003 May 30. Mol Biol Evol. 2003. PMID: 12777511 - Alu repeats and human genomic diversity.
Batzer MA, Deininger PL. Batzer MA, et al. Nat Rev Genet. 2002 May;3(5):370-9. doi: 10.1038/nrg798. Nat Rev Genet. 2002. PMID: 11988762 Review. - Alu elements and the human genome.
Rowold DJ, Herrera RJ. Rowold DJ, et al. Genetica. 2000;108(1):57-72. doi: 10.1023/a:1004099605261. Genetica. 2000. PMID: 11145422 Review.
Cited by
- Biosynthesis of Circular RNA ciRS-7/CDR1as Is Mediated by Mammalian-wide Interspersed Repeats.
Yoshimoto R, Rahimi K, Hansen TB, Kjems J, Mayeda A. Yoshimoto R, et al. iScience. 2020 Jul 24;23(7):101345. doi: 10.1016/j.isci.2020.101345. Epub 2020 Jul 4. iScience. 2020. PMID: 32683316 Free PMC article. - RNA transcription and degradation of Alu retrotransposons depends on sequence features and evolutionary history.
Baar T, Dümcke S, Gressel S, Schwalb B, Dilthey A, Cramer P, Tresch A. Baar T, et al. G3 (Bethesda). 2022 May 6;12(5):jkac054. doi: 10.1093/g3journal/jkac054. G3 (Bethesda). 2022. PMID: 35253846 Free PMC article. - Liver X Receptor-Binding DNA Motif Associated With Atherosclerosis-Specific DNA Methylation Profiles of Alu Elements and Neighboring CpG Islands.
Tristán-Flores FE, Guzmán P, Ortega-Kermedy MS, Cruz-Torres G, de la Rocha C, Silva-Martínez GA, Rodríguez-Ríos D, Alvarado-Caudillo Y, Barbosa-Sabanero G, Sayols S, Lund G, Zaina S. Tristán-Flores FE, et al. J Am Heart Assoc. 2018 Jan 31;7(3):e007686. doi: 10.1161/JAHA.117.007686. J Am Heart Assoc. 2018. PMID: 29386205 Free PMC article. - Alu repeat discovery and characterization within human genomes.
Hormozdiari F, Alkan C, Ventura M, Hajirasouliha I, Malig M, Hach F, Yorukoglu D, Dao P, Bakhshi M, Sahinalp SC, Eichler EE. Hormozdiari F, et al. Genome Res. 2011 Jun;21(6):840-9. doi: 10.1101/gr.115956.110. Epub 2010 Dec 3. Genome Res. 2011. PMID: 21131385 Free PMC article. - Identification of three new Alu Yb subfamilies by source tracking of recently integrated Alu Yb elements.
Ahmed M, Li W, Liang P. Ahmed M, et al. Mob DNA. 2013 Nov 12;4(1):25. doi: 10.1186/1759-8753-4-25. Mob DNA. 2013. PMID: 24216009 Free PMC article.
References
- Arndt, P.F., Petrov, D.A., and Hwa, T. 2003. Distinct changes of genomic biases in nucleotide substitution at the time of mammalian radiation. Mol. Biol. Evol. 20: 1887-1896. - PubMed
- Batzer, M.A. and Deininger, P.L. 1991. A human-specific subfamily of Alu sequences. Genomics 9: 481-487. - PubMed
- ———. 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3: 370-379. - PubMed
WEB SITE REFERENCES
- http://repeatmasker.org; RepeatMasker.
- http://www.cs.ucsd.edu/~aprice/alu.html; implementation of our algorithm.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources