New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria - PubMed (original) (raw)
New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria
Maryvonne Arnaud et al. Appl Environ Microbiol. 2004 Nov.
Abstract
A shuttle vector designated pMAD was constructed for quickly generating gene inactivation mutants in naturally nontransformable gram-positive bacteria. This vector allows, on X-Gal (5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside) plates, a quick colorimetric blue-white discrimination of bacteria which have lost the plasmid, greatly facilitating clone identification during mutagenesis. The plasmid was used in Staphylococcus aureus, Listeria monocytogenes, and Bacillus cereus to efficiently construct mutants with or without an associated antibiotic resistance gene.
Figures
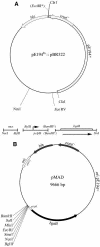
FIG. 1.
Physical map of pE194ts::pBR322 and its derivative pMAD. (A) Construction of the pMAD plasmid. DNA fragments used to construct pMAD are indicated: mcs, multiple cloning site; pclpB, DNA fragment from S. aureus containing the clpB promoter region; bgaB, DNA fragment containing the promoterless bgaB gene encoding a thermostable β-galactosidase (8). Asterisks indicate restriction sites which have been lost during the construction steps. (B) Map of the pMAD plasmid. Unique restriction sites are indicated.
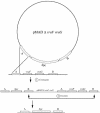
FIG. 2.
Schematic representation of a two-step procedure used to obtain gene replacement recombination. Areas labeled A and B represent DNA sequences located upstream and downstream from vraF and vraG genes. The crossed lines indicate crossover events. The integration of pMAD via homologous sequences can take place in area A or B. The cointegrate undergoes a second recombination event, regenerating the pMAD plasmid. Depending on whether the second recombination event occurs between the two homologous sequences in area A or B, the spectinomycin resistance marker will either remain in the chromosome (area B) or be excised along with the plasmid (area A). Gene replacement occurs only if the second recombination event occurs in area B, as shown.
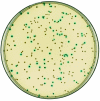
FIG. 3.
L. monocytogenes LO28 colonies following transformation with the pMADΔ_clpB_ plasmid. Cells were cultivated at 30°C for 6 h, followed by incubation at 39°C for 3 h; dilutions were plated on TSA X-Gal plates in the absence of antibiotics. White colonies have undergone the excision and loss of the plasmid vector, whereas blue colonies retain a copy of the plasmid integrated in the chromosome.
Similar articles
- A chimeric vector for efficient chromosomal modification in Enterococcus faecalis and other lactic acid bacteria.
Blancato VS, Magni C. Blancato VS, et al. Lett Appl Microbiol. 2010 May;50(5):542-6. doi: 10.1111/j.1472-765X.2010.02815.x. Epub 2010 Feb 1. Lett Appl Microbiol. 2010. PMID: 20158606 - Transformation of microorganisms with the plasmid vector with the replicon from pAC1 from Acetobacter pasteurianus.
Grones J, Turna J. Grones J, et al. Biochem Biophys Res Commun. 1995 Jan 26;206(3):942-7. doi: 10.1006/bbrc.1995.1133. Biochem Biophys Res Commun. 1995. PMID: 7832808 - [A family of shuttle vectors for lactobacilli and other gram-positive bacteria based on the plasmid pLF1311 replicon].
Aleshin VV, Semenova EV, Tarakanov BV, Livshits VA. Aleshin VV, et al. Mikrobiologiia. 2000 Jan-Feb;69(1):75-80. Mikrobiologiia. 2000. PMID: 10808493 Russian. - The role of sigmaB in the stress response of Gram-positive bacteria -- targets for food preservation and safety.
van Schaik W, Abee T. van Schaik W, et al. Curr Opin Biotechnol. 2005 Apr;16(2):218-24. doi: 10.1016/j.copbio.2005.01.008. Curr Opin Biotechnol. 2005. PMID: 15831390 Review. - A modular master on the move: the Tn916 family of mobile genetic elements.
Roberts AP, Mullany P. Roberts AP, et al. Trends Microbiol. 2009 Jun;17(6):251-8. doi: 10.1016/j.tim.2009.03.002. Epub 2009 May 20. Trends Microbiol. 2009. PMID: 19464182 Review.
Cited by
- The megaplasmid pCER270 of Bacillus cereus emetic strain affects the timing of the sporulation process, spore resistance properties, and germination.
Perchat S, Nevers A, Kranzler M, Ehling-Schulz M, Lereclus D, Gohar M. Perchat S, et al. Appl Environ Microbiol. 2024 Sep 18;90(9):e0102924. doi: 10.1128/aem.01029-24. Epub 2024 Aug 19. Appl Environ Microbiol. 2024. PMID: 39158315 - Membrane Localization of RNase Y Is Important for Global Gene Expression in Bacillus subtilis.
Laalami S, Cavaiuolo M, Oberto J, Putzer H. Laalami S, et al. Int J Mol Sci. 2024 Aug 5;25(15):8537. doi: 10.3390/ijms25158537. Int J Mol Sci. 2024. PMID: 39126106 Free PMC article. - ABD-3, the confluence of powerful antibacterial modalities: ABDs delivering and expressing _ls_s, the gene encoding lysostaphin.
Ram G, Chiu L, Dey S, Ross HF, Cammer M, Novick RP. Ram G, et al. Antimicrob Agents Chemother. 2024 Sep 4;68(9):e0023524. doi: 10.1128/aac.00235-24. Epub 2024 Jul 29. Antimicrob Agents Chemother. 2024. PMID: 39072634 - Genetic Engineering of Bacillus subtilis Using Competence-Induced Homologous Recombination Techniques.
Meijer WJJ, Miguel-Arribas A. Meijer WJJ, et al. Methods Mol Biol. 2024;2819:241-260. doi: 10.1007/978-1-0716-3930-6_12. Methods Mol Biol. 2024. PMID: 39028510 - Flotillin-mediated stabilization of unfolded proteins in bacterial membrane microdomains.
Ukleja M, Kricks L, Torrens G, Peschiera I, Rodrigues-Lopes I, Krupka M, García-Fernández J, Melero R, Del Campo R, Eulalio A, Mateus A, López-Bravo M, Rico AI, Cava F, Lopez D. Ukleja M, et al. Nat Commun. 2024 Jul 3;15(1):5583. doi: 10.1038/s41467-024-49951-1. Nat Commun. 2024. PMID: 38961085 Free PMC article.
References
- Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. - PubMed
- Chambers, S. P., S. E. Prior, D. A. Barstow, and N. P. Minton. 1988. The pMTL nic− cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene 68:139-149. - PubMed
- Chastanet, A., J. Fert, and T. Msadek. 2003. Comparative genomics reveal novel heat-shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol. Microbiol. 47:1061-1073. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous