Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome - PubMed (original) (raw)
Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome
Alekos Athanasiadis et al. PLoS Biol. 2004 Dec.
Abstract
RNA editing by adenosine deamination generates RNA and protein diversity through the posttranscriptional modification of single nucleotides in RNA sequences. Few mammalian A-to-I edited genes have been identified despite evidence that many more should exist. Here we identify intramolecular pairs of Alu elements as a major target for editing in the human transcriptome. An experimental demonstration in 43 genes was extended by a broader computational analysis of more than 100,000 human mRNAs. We find that 1,445 human mRNAs (1.4%) are subject to RNA editing at more than 14,500 sites, and our data further suggest that the vast majority of pre-mRNAs (greater than 85%) are targeted in introns by the editing machinery. The editing levels of Alu-containing mRNAs correlate with distance and homology between inverted repeats and vary in different tissues. Alu-mediated RNA duplexes targeted by RNA editing are formed intramolecularly, whereas editing due to intermolecular base-pairing appears to be negligible. We present evidence that these editing events can lead to the posttranscriptional creation or elimination of splice signals affecting alternatively spliced Alu-derived exons. The analysis suggests that modification of repetitive elements is a predominant activity for RNA editing with significant implications for cellular gene expression.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures
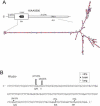
Figure 1. RNA Editing of an Alternatively Spliced Alu-Exon in a G-Protein Coupled Receptor
(A) Schematic representation of LUSTR (GPR107, KIAA1624) gene structure around edited exon 15a. The AluSx repeat element in intron 15 and the exonic, inversely oriented AluJo are predicted to form an intramolecular foldback structure as depicted below (MFold software). TM, exonic regions predicted to encode transmembrane domains; *, editing sites. (B) Editing analysis of exon 15a (sequence in capital letters) and flanking regions. The two major editing sites predicted to change amino acids (H/R and Q/R) are indicated. Editing levels in brain (filled column) and lung (open column) are shown above each edited nucleotide. The splice acceptor site subject to editing is underlined.
Figure 3. RNA Editing of Alternative Exon 22a in Inhibitor BTKI and the 5′-UTR of KIAA1497
(A) The alternatively spliced exon 22a and surrounding region of the BTKI (KIAA1417) gene with two Alu elements and its computer-predicted foldback structure. (B) Editing analysis of the AluSx- element with the exonic sequence in capital letters and edited A's in bold. The alternative splice acceptor site is underlined with a dashed line; the additional alternative consensus splice acceptor site, which undergoes editing, is underlined with a solid line. (C) Gene architecture and Alu foldback structure of KIAA1497. The brain-derived cDNA of KIAA1497, also known as LRRN1; (Taguchi et al. 1996), has a total of 15 nonpolymorphic AtoG discrepancies to gDNA, 14 being located within the 5′-UTR of the gene and one within the coding region. We analyzed PCR products covering all 14 potential editing sites in the 5′-UTR for editing in cDNA from human brain and could confirm in vivo editing to an extend clearly above the detection limit of our method for most of these positions and also at additional adenosines (data not shown). ORF, open reading frame; *, editing sites.
Figure 2. RNA Editing of KIAA500 Alu Inverted Repeat
KIAA0500 is a cDNA of 6,577 nt in length cloned from human brain (AB007969) with a predicted open reading frame of 213 amino acids. Four AtoG discrepancies were present within the coding region of which two lead to an amino acid change (Q/R and S/G, respectively). (A) Structure of the KIAA500 mRNA with location of Alu elements indicated and the predicted RNA secondary structure according to the MFOLD algorithm. Large open box indicates predicted open reading frame. *, editing sites. (B) Editing analysis of an exonic Alu element in KIAA0500. Editing sites predicted to change amino acids are indicated. Our analysis revealed a significant percentage of editing (%G) at the nucleotide positions 3518 (27% ± 3%), 3522 (20% ± 3%) and 3625 (6% ± 1%) and additional editing sites with less than 5% editing, whereas parallel analysis of human gDNA confirmed the presence of adenosine at these positions. Editing levels in brain (filled column) and lung (open column, where detectable) are shown above each edited nucleotide.
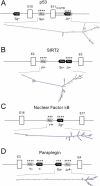
Figure 4. Alu-Mediated RNA Editing in p53, SIRT2, NFκB, and Paraplegin Pre-mRNAs
Schematic presentation of the gene structures from (A) P53, (B) SIRT2, (C) NFκB, and (D) SPG7. Edited repeat elements are marked by asterisks. RNA folds appear as calculated with MFOLD. The AluJb− in p53 is located in the 3′-UTR (A); all others are intronic. *, editing sites.
Figure 5. Editing of Alternative Foldback Structures of GPR81 Pre-mRNA
(A) The position and orientation of all four Alu elements in GPR81 pre-mRNA is indicated. Three alternative Alu pairings (I–III) are predicted and experimental editing analysis indicates that all three do form in vivo. ORF, open reading frame; *, editing sites. (B) Editing analysis of AluSp+ in GPR81. Percentages of editing in human brain are indicated. The exonic sequence appears in capitals. The edited AT dinucleotide that becomes a splice donor site is underlined.
Figure 6. Mismatch Bias in Exonic Repetitive-Element Sequences
(A) Plot of the nature and number of mismatches within Alu and L1 sequences present in human cDNAs. For reasons of comparison the L1 mismatch numbers have been multiplied by 2.9 so that the non-AtoG mismatch count for Alu and L1 is identical. Transition mismatches AtoG, GtoA, CtoT, and TtoC are displayed together for comparison. (B) Plotted are the total number of Alu sequences found in human cDNAs (first column) and the number of elements harboring AtoG and GtoA mismatches (second and last column). The third column indicates the high confidence set of edited elements (α = 0.000001).
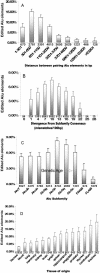
Figure 7. Factors Determining Alu Targeting Probability
(A) Percentage of edited elements classified in bins according to the distance separating the element and its closest inverted Alu partner. (B) Percentage of edited Alu elements clustered according to divergence from their corresponding Alu-subfamily consensus. (C) Percentage of edited Alu elements in each Alu subfamily. In (A), (B), and (C) the numbers at the bottom of the bars show the sample size in each bin. (D) Percentage of edited elements according to the tissue from which the RNA was isolated. Error bars show 95% confidence levels.
Figure 8. Sequence and Structure Preferences of Editing in Alus
(A) The consensus sequence of 141 edited full-length Alu elements present within human Chromosome 1 transcripts with the number of editing events indicated for each sequence position (bars). Insertions and deletions present in fewer than five elements are not shown in the alignment for clarity. Bases conserved in more than 80% of the sequences are boxed. For the lesser conserved consensus positions the next most frequent base is listed below. Consensus CpG dinucleotides are in bold. Arrows indicate “high-efficiency” positions where more than 20% of adenosines present appear to be edited. Note the overlap of these positions with CpGs. Major features of Alu sequences, such as the A-Box and B-Box of Pol III and the Alu polyA sequence are labeled. (B) A typical Alu foldback structure and its major features as discussed in the text. Arrows indicate TA hot-spot positions. The magnifications show the two typical configurations of editing sites found in Alu pairs: mismatched A/C bulges (i) and A/U base pairs (ii).
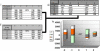
Figure 9. Cis Preferences of Editing Sites in Alus
Tables (i) and (ii) show the frequency of A, G, C, T, or an A/G editing site at positions −2, −1, 1, and 2 relative to each of the 14,774 AtoG mismatch sites found within the high confidence group of Alu elements (i) and in relation to a randomly chosen adenosine from each of the those sequences for each AtoG mismatch (ii). Table (iii) shows relative editing preferences after bias removal by subtracting table (ii) from Table (i). (iv) Graphical representation of Table (iii).
Similar articles
- Noncoding regions of C. elegans mRNA undergo selective adenosine to inosine deamination and contain a small number of editing sites per transcript.
Wheeler EC, Washburn MC, Major F, Rusch DB, Hundley HA. Wheeler EC, et al. RNA Biol. 2015;12(2):162-74. doi: 10.1080/15476286.2015.1017220. RNA Biol. 2015. PMID: 25826568 Free PMC article. - Identification of widespread ultra-edited human RNAs.
Carmi S, Borukhov I, Levanon EY. Carmi S, et al. PLoS Genet. 2011 Oct;7(10):e1002317. doi: 10.1371/journal.pgen.1002317. Epub 2011 Oct 20. PLoS Genet. 2011. PMID: 22028664 Free PMC article. - Widespread RNA editing of embedded alu elements in the human transcriptome.
Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Kim DD, et al. Genome Res. 2004 Sep;14(9):1719-25. doi: 10.1101/gr.2855504. Genome Res. 2004. PMID: 15342557 Free PMC article. - Editor meets silencer: crosstalk between RNA editing and RNA interference.
Nishikura K. Nishikura K. Nat Rev Mol Cell Biol. 2006 Dec;7(12):919-31. doi: 10.1038/nrm2061. Nat Rev Mol Cell Biol. 2006. PMID: 17139332 Free PMC article. Review. - A-to-I editing of coding and non-coding RNAs by ADARs.
Nishikura K. Nishikura K. Nat Rev Mol Cell Biol. 2016 Feb;17(2):83-96. doi: 10.1038/nrm.2015.4. Epub 2015 Dec 9. Nat Rev Mol Cell Biol. 2016. PMID: 26648264 Free PMC article. Review.
Cited by
- Noncoding regions of C. elegans mRNA undergo selective adenosine to inosine deamination and contain a small number of editing sites per transcript.
Wheeler EC, Washburn MC, Major F, Rusch DB, Hundley HA. Wheeler EC, et al. RNA Biol. 2015;12(2):162-74. doi: 10.1080/15476286.2015.1017220. RNA Biol. 2015. PMID: 25826568 Free PMC article. - Inverted repeat Alu elements in the human lincRNA-p21 adopt a conserved secondary structure that regulates RNA function.
Chillón I, Pyle AM. Chillón I, et al. Nucleic Acids Res. 2016 Nov 2;44(19):9462-9471. doi: 10.1093/nar/gkw599. Epub 2016 Jul 4. Nucleic Acids Res. 2016. PMID: 27378782 Free PMC article. - Impact of retrotransposons in pluripotent stem cells.
Tanaka Y, Chung L, Park IH. Tanaka Y, et al. Mol Cells. 2012 Dec;34(6):509-16. doi: 10.1007/s10059-012-0242-8. Epub 2012 Nov 6. Mol Cells. 2012. PMID: 23135636 Free PMC article. Review. - Antiviral Defence Mechanisms during Early Mammalian Development.
Mueller F, Witteveldt J, Macias S. Mueller F, et al. Viruses. 2024 Jan 24;16(2):173. doi: 10.3390/v16020173. Viruses. 2024. PMID: 38399949 Free PMC article. Review. - The human transcriptome: an unfinished story.
Pertea M. Pertea M. Genes (Basel). 2012 Sep;3(3):344-60. doi: 10.3390/genes3030344. Genes (Basel). 2012. PMID: 22916334 Free PMC article.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, et al. Current protocols in molecular biology (vols. 1–4) New York: Wiley; 1995.
- Baltimore D. Our genome unveiled. Nature. 2001;409:814–816. - PubMed
- Bass BL, Weintraub H. A developmentally regulated activity that unwinds RNA duplexes. Cell. 1987;48:607–613. - PubMed
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources