The E2F family: specific functions and overlapping interests - PubMed (original) (raw)
Review
The E2F family: specific functions and overlapping interests
Claire Attwooll et al. EMBO J. 2004.
Abstract
The E2F transcription factors are key regulators of cell cycle progression and the E2F field has made rapid advances since its advent in 1986. Yet, while our understanding of the roles and functions of the E2F family has made enormous progress, with each discovery new questions arise. In this review, we summarise the most recent advances in the field and discuss the remaining key questions. In particular, we will focus on how specificity is achieved among the E2Fs.
Figures
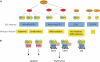
Figure 1
Schematic representation of the E2F transcription factor subgroups, their physiological roles and specific binding partners. (A) The E2F family is divided into at least four subgroups defined by their regulation by pocket proteins and chromatin modifiers, and by their physiological function (i.e. activation or repression). See text for details. ‘DP' presents DP1 or DP2, ‘chromatin modifiers' refer to chromatin-modifying activities binding to the pRB family members, such as HDAC, Suv39H, BRG1 or HPC2. Recent results from our laboratory have shown that also E2F7 recruits repressors to E2F-dependent promoters (Di Stefano and Helin, unpublished results). (B) Specificity among the E2Fs through protein–protein interactions. TopBP1 and GABPγ1 are specific binding partners of E2F1, while TFE3 binds only to E2F3, and RYBP and YY1 bind only to E2Fs 2 and 3.
Figure 2
Models for the regulation of gene expression by the E2F transcription factors. (A) Classical model for the regulation of E2F-dependent promoters by the E2Fs. This model is based on the original observations of E2F-binding partners, localisation studies and promoter affinities. See text for details of this model. (B) The ‘ChIP model' for E2F regulation of target promoters. Using ChIP assays, pRB has not been found to be associated with promoters containing the E2F consensus site during normal cell cycle progression. Thus, the ChIP model predicts that p130- and p107-, but not pRB-containing complexes, regulate E2F-dependent transcription in proliferating cells, whereas pRB-containing complexes may have a role in regulating E2F-dependent promoters in cells exiting the cell cycle (senescence/terminal differentiation). In addition to HDAC, chromatin modifiers such as Suv39H, BRG1 and HPC2 have also been shown to associate with pRB. (C) The ‘combined model' for E2F regulation of target promoters. This model combines those shown in panels A and B, and postulates that pRB, in association with E2F-DP, also regulates the expression of thus far unidentified E2F target genes (‘non-ChIP targets'), which are important for the regulation of the cell cycle. This model is supported by the observation that inactivation of pRb in mouse cells lead to deregulation of the cell cycle.
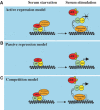
Figure 3
Models for the mechanism of regulation of E2F-dependent promoters. (A) ‘Active repression model': repressing E2Fs recruit chromatin modifiers to E2F-dependent promoters; thus, this repression must be relieved (chromatin remodelling) before promoters can be activated. In addition, the repressing E2Fs physically prevent binding of the E2F target promoter by the activating E2Fs. In this model, and in the models shown in panels B and C, pRB represents the pRB family members. (B) ‘Passive repression model': pRB prevents E2F promoter activation by binding to the transactivation domains of the E2Fs. This binding may also prevent the recruitment of essential co-activators to the promoter. Repression is relieved by phosphorylation of pRB and its concomitant dissociation from E2Fs, freeing the transactivation domain for activity. (C) ‘Competition model': the activating and repressing E2Fs compete for binding to the E2F DNA-binding site. This model also includes the recruitment of chromatin modifiers by pRB family members.
Similar articles
- Hitting their targets: an emerging picture of E2F and cell cycle control.
Blais A, Dynlacht BD. Blais A, et al. Curr Opin Genet Dev. 2004 Oct;14(5):527-32. doi: 10.1016/j.gde.2004.07.003. Curr Opin Genet Dev. 2004. PMID: 15380244 Review. - Molecular mechanisms of E2F-dependent activation and pRB-mediated repression.
Frolov MV, Dyson NJ. Frolov MV, et al. J Cell Sci. 2004 May 1;117(Pt 11):2173-81. doi: 10.1242/jcs.01227. J Cell Sci. 2004. PMID: 15126619 Review. - E2Fs link the control of G1/S and G2/M transcription.
Zhu W, Giangrande PH, Nevins JR. Zhu W, et al. EMBO J. 2004 Nov 24;23(23):4615-26. doi: 10.1038/sj.emboj.7600459. Epub 2004 Oct 28. EMBO J. 2004. PMID: 15510213 Free PMC article. - Evolving intricacies and implications of E2F1 regulation.
Mundle SD, Saberwal G. Mundle SD, et al. FASEB J. 2003 Apr;17(6):569-74. doi: 10.1096/fj.02-0431rev. FASEB J. 2003. PMID: 12665469 Review. - Endocycle and E2F-dependent transcriptional activation and repression.
Du W. Du W. Cell Cycle. 2003 Nov-Dec;2(6):515-6. doi: 10.4161/cc.2.6.579. Cell Cycle. 2003. PMID: 14504463 No abstract available.
Cited by
- FBXO5-mediated RNF183 degradation prevents endoplasmic reticulum stress-induced apoptosis and promotes colon cancer progression.
Ji J, Jing A, Ding Y, Ma X, Qian Q, Geng T, Cheng W, Zhang M, Sun Q, Ma S, Wang X, Yuan Q, Xu M, Qin J, Ma L, Yang J, He J, Du Q, Xia M, Xu Y, Chen Z, Zhu L, Liu W, Liu S, Liu B. Ji J, et al. Cell Death Dis. 2024 Jan 11;15(1):33. doi: 10.1038/s41419-024-06421-2. Cell Death Dis. 2024. PMID: 38212299 Free PMC article. - Comprehensive bioinformatics analysis of the E2F family in human clear cell renal cell carcinoma.
Liu ZG, Su J, Liu H, Yang XJ, Yang X, Wei Y, Zhu XY, Song Y, Zhao XC, Guo HL. Liu ZG, et al. Oncol Lett. 2022 Aug 19;24(4):351. doi: 10.3892/ol.2022.13471. eCollection 2022 Oct. Oncol Lett. 2022. PMID: 36168311 Free PMC article. - DNM3OS Enhances the Apoptosis and Senescence of Spermatogonia Associated with Nonobstructive Azoospermia by Providing miR-214-5p and Decreasing E2F2 Expression.
Hua R, Chu Q, Guo F, Chen Q, Li M, Zhou X, Zhu Y. Hua R, et al. Anal Cell Pathol (Amst). 2023 Dec 20;2023:1477658. doi: 10.1155/2023/1477658. eCollection 2023. Anal Cell Pathol (Amst). 2023. PMID: 38152068 Free PMC article. - E2F1 copy number variations in germline and breast cancer: a retrospective study of 222 Italian women.
Rocca MS, Benna C, Goldin E, Di Nisio A, De Toni L, Cosci I, Marchet A, Nitti D, Foresta C. Rocca MS, et al. Mol Med. 2021 Mar 10;27(1):26. doi: 10.1186/s10020-021-00287-2. Mol Med. 2021. PMID: 33691613 Free PMC article. - Overexpression Populus d-Type Cyclin Gene PsnCYCD1;1 Influences Cell Division and Produces Curved Leaf in Arabidopsis thaliana.
Zheng T, Dai L, Liu Y, Li S, Zheng M, Zhao Z, Qu GZ. Zheng T, et al. Int J Mol Sci. 2021 May 29;22(11):5837. doi: 10.3390/ijms22115837. Int J Mol Sci. 2021. PMID: 34072501 Free PMC article.
References
- Cartwright P, Müller H, Wagener C, Holm K, Helin K (1998) E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene 17: 611–624 - PubMed
- Chellappan S, Hiebert S, Mudryj M, Horowitz J, Nevins J (1991) The E2F transcription factor is a cellular target for the RB protein. Cell 65: 1053–1061 - PubMed
- Croxton R, Ma Y, Song L, Haura EB, Cress WD (2002) Direct repression of the Mcl-1 promoter by E2F1. Oncogene 21: 1359–1369 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases