Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips - PubMed (original) (raw)
Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips
Jing Huang et al. Proc Natl Acad Sci U S A. 2004.
Abstract
The TOR (target of rapamycin) proteins play important roles in nutrient signaling in eukaryotic cells. Rapamycin treatment induces a state reminiscent of the nutrient starvation response, often resulting in growth inhibition. Using a chemical genetic modifier screen, we identified two classes of small molecules, small-molecule inhibitors of rapamycin (SMIRs) and small-molecule enhancers of rapamycin (SMERs), that suppress and augment, respectively, rapamycin's effect in the yeast Saccharomyces cerevisiae. Probing proteome chips with biotinylated SMIRs revealed putative intracellular target proteins, including Tep1p, a homolog of the mammalian PTEN (phosphatase and tensin homologue deleted on chromosome 10) tumor suppressor, and Ybr077cp (Nir1p), a protein of previously unknown function that we show to be a component of the TOR signaling network. Both SMIR target proteins are associated with PI(3,4)P2, suggesting a mechanism of regulation of the TOR pathway involving phosphatidylinositides. Our results illustrate the combined use of chemical genetics and proteomics in biological discovery and map a path for creating useful therapeutics for treating human diseases involving the TOR pathway, such as diabetes and cancer.
Figures
Fig. 1.
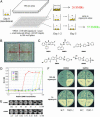
A chemical genetic screen for small molecules that modulate rapamycin's antiproliferative effect in yeast. (A) Schematics of the screen. Compounds were transferred from library plates to assay plates (containing growth medium, rapamycin, and yeast cells) by using 384-pin arrays. SMER, small-molecule enhancers of rapamycin. (B) Retest of SMIRs in a 384-well plate. White wells indicate compound-induced yeast growth in the presence of rapamycin; black (transparent) wells indicate no growth. (C) Chemical structures of the fast-acting SMIRs (yeast growth identifiable on day 1, same as the “no rapamycin” control). (D) Dose–response curves for SMIRs in wild-type (rapamycin-sensitive EGY48) cells inoculated in YPDA containing 100 nM rapamycin. (E) Minimal concentrations of SMIR3 and SMIR4 required for yeast growth in YPDA containing 20 nM rapamycin. (F) SMIR4 treatment and the _TOR1_-1 (S1972R) mutation (26) both confer rapamycin resistance. Cells were plated at two different densities on the upper versus lower halves of the plates (1:1,000).
Fig. 2.
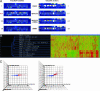
SMIRs suppress rapamycin's effect in yeast on the whole-genome scale as revealed by mRNA transcript abundance analysis. (A) Example views of GeneChip features containing probes (outlined in white) specific for the GDH3 and URA7 transcripts (up- and down-regulated by rapamycin treatment, respectively) on the Affymetrix Ye6100 oligonucleotide arrays. (B) For comparing the effects of SMIRs and genetic mutations, a 2D clustergram with experiment tree and gene tree was generated in
genespring
(Silicon Genetics, Redwood City, CA) by using the hierarchical clustering program (minimal distance, 0.001; separation ratio, 5; new tree with standard correlation). (C) Profiling data visualized by 3D scatter plots in
genespring
. Fold changes over DMSO mock treatment were plotted for 6,430 genes on the Affymetrix chip. Color scaling is based on the rapamycin profile: red, increase; blue, decrease.
Fig. 3.
Protein targets of SMIR4 identified by binding of SMIR4-biotin to the yeast proteome chip (15). (A)(Left) Image of the whole proteome chip. (Right) Enlarged images of the areas that contain the positive twin-spots. Four proteins that bind strongly to SMIR4 are indicated. (B and C) Yeast cell growth on agar. (B) Ybr077cp, one of the SMIR4-binding proteins identified in A, when deleted causes hypersensitivity to rapamycin and insensitivity to the suppressive effect of SMIR4, indicating that Ybr077cp is a bona fide target of SMIR4. (C) Ybr077cp is also required for SMIR3 function, suggesting possible common mechanisms by SMIR3 and SMIR4.
Fig. 4.
Ybr077cp (Nir1p) is a new component of the TOR signaling network. (A) Model of SMIR3 and SMIR4 suppression of the antiproliferative effect of rapamycin possibly by modulating PIs and thereby regulating Ybr077cp activity. (B) Graphic representation of a Ybr077cp protein interaction network and the rapamycin response phenotypes of individual deletion strains. Red, rapamycin-resistant when deleted; green, rapamycin-hypersensitive when deleted; yellow, essential genes whose heterozygous deletion did not produce detectable change in rapamycin sensitivity. (C) Gene-expression evidence for NIR1 function in the Tor network. NC, no change; D, decrease.
Similar articles
- TORC1 controls degradation of the transcription factor Stp1, a key effector of the SPS amino-acid-sensing pathway in Saccharomyces cerevisiae.
Shin CS, Kim SY, Huh WK. Shin CS, et al. J Cell Sci. 2009 Jun 15;122(Pt 12):2089-99. doi: 10.1242/jcs.047191. J Cell Sci. 2009. PMID: 19494127 - Hsf1 activation inhibits rapamycin resistance and TOR signaling in yeast revealed by combined proteomic and genetic analysis.
Bandhakavi S, Xie H, O'Callaghan B, Sakurai H, Kim DH, Griffin TJ. Bandhakavi S, et al. PLoS One. 2008 Feb 13;3(2):e1598. doi: 10.1371/journal.pone.0001598. PLoS One. 2008. PMID: 18270585 Free PMC article. - Identification of Novel Components of Target-of-Rapamycin Signaling Pathway by Network-Based Multi-Omics Integrative Analysis.
Dereli Eke E, Arga KY, Dikicioglu D, Eraslan S, Erkol E, Celik A, Kirdar B, Di Camillo B. Dereli Eke E, et al. OMICS. 2019 May;23(5):274-284. doi: 10.1089/omi.2019.0021. Epub 2019 Apr 13. OMICS. 2019. PMID: 30985253 - Control of translation by the target of rapamycin proteins.
Gingras AC, Raught B, Sonenberg N. Gingras AC, et al. Prog Mol Subcell Biol. 2001;27:143-74. doi: 10.1007/978-3-662-09889-9_6. Prog Mol Subcell Biol. 2001. PMID: 11575159 Review. No abstract available. - Halcyon days of TOR: Reflections on the multiple independent discovery of the yeast and mammalian TOR proteins.
Livi GP. Livi GP. Gene. 2019 Apr 15;692:145-155. doi: 10.1016/j.gene.2018.12.046. Epub 2019 Jan 9. Gene. 2019. PMID: 30639424 Review.
Cited by
- Turning liabilities into opportunities: Off-target based drug repurposing in cancer.
Palve V, Liao Y, Remsing Rix LL, Rix U. Palve V, et al. Semin Cancer Biol. 2021 Jan;68:209-229. doi: 10.1016/j.semcancer.2020.02.003. Epub 2020 Feb 7. Semin Cancer Biol. 2021. PMID: 32044472 Free PMC article. Review. - Changes of Cell Biochemical States Are Revealed in Protein Homomeric Complex Dynamics.
Stynen B, Abd-Rabbo D, Kowarzyk J, Miller-Fleming L, Aulakh SK, Garneau P, Ralser M, Michnick SW. Stynen B, et al. Cell. 2018 Nov 15;175(5):1418-1429.e9. doi: 10.1016/j.cell.2018.09.050. Epub 2018 Oct 25. Cell. 2018. PMID: 30454649 Free PMC article. - The Potentials and Pitfalls of Microarrays in Neglected Tropical Diseases: A Focus on Human Filarial Infections.
Kwarteng A, Ahuno ST. Kwarteng A, et al. Microarrays (Basel). 2016 Aug 2;5(3):20. doi: 10.3390/microarrays5030020. Microarrays (Basel). 2016. PMID: 27600086 Free PMC article. Review. - Applications of protein biochips in biomedical and biotechnological research.
Weinrich D, Jonkheijm P, Niemeyer CM, Waldmann H. Weinrich D, et al. Angew Chem Int Ed Engl. 2009;48(42):7744-51. doi: 10.1002/anie.200901480. Angew Chem Int Ed Engl. 2009. PMID: 19757463 Free PMC article. Review. - Global screening of genes essential for growth in high-pressure and cold environments: searching for basic adaptive strategies using a yeast deletion library.
Abe F, Minegishi H. Abe F, et al. Genetics. 2008 Feb;178(2):851-72. doi: 10.1534/genetics.107.083063. Epub 2008 Feb 1. Genetics. 2008. PMID: 18245339 Free PMC article.
References
- DeRisi, J. L., Iyer, V. R. & Brown, P. O. (1997) Science 278, 680–686. - PubMed
- Hartwell, L. H., Hopfield, J. J., Leibler, S. & Murray, A. W. (1999) Nature 402, C47–C52. - PubMed
- Hughes, T. R., Marton, M. J., Jones, A. R., Roberts, C. J., Stoughton, R., Armour, C. D., Bennett, H. A., Coffey, E., Dai, H., He, Y. D., et al. (2000) Cell 102, 109–126. - PubMed
- Bishop, A. C., Ubersax, J. A., Petsch, D. T., Matheos, D. P., Gray, N. S., Blethrow, J., Shimizu, E., Tsien, J. Z., Schultz, P. G., Rose, M. D., et al. (2000) Nature 407, 395–401. - PubMed
- Ideker, T., Thorsson, V., Ranish, J. A., Christmas, R., Buhler, J., Eng, J. K., Bumgarner, R., Goodlett, D. R., Aebersold, R. & Hood, L. (2001) Science 292, 929–934. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous