Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays - PubMed (original) (raw)
Comparative Study
. 2004 Dec;36(12):1331-9.
doi: 10.1038/ng1473. Epub 2004 Nov 14.
Affiliations
- PMID: 15543148
- PMCID: PMC2692596
- DOI: 10.1038/ng1473
Comparative Study
Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays
Sonali Mukherjee et al. Nat Genet. 2004 Dec.
Abstract
We developed a new DNA microarray-based technology, called protein binding microarrays (PBMs), that allows rapid, high-throughput characterization of the in vitro DNA binding-site sequence specificities of transcription factors in a single day. Using PBMs, we identified the DNA binding-site sequence specificities of the yeast transcription factors Abf1, Rap1 and Mig1. Comparison of these proteins' in vitro binding sites with their in vivo binding sites indicates that PBM-derived sequence specificities can accurately reflect in vivo DNA sequence specificities. In addition to previously identified targets, Abf1, Rap1 and Mig1 bound to 107, 90 and 75 putative new target intergenic regions, respectively, many of which were upstream of previously uncharacterized open reading frames. Comparative sequence analysis indicated that many of these newly identified sites are highly conserved across five sequenced sensu stricto yeast species and, therefore, are probably functional in vivo binding sites that may be used in a condition-specific manner. Similar PBM experiments should be useful in identifying new cis regulatory elements and transcriptional regulatory networks in various genomes.
Figures
Figure 1. Protein binding microarray (PBM) schematic
(a) Overview of PBM experiments. (b) Whole-genome yeast intergenic microarray bound by Rap1. The fluorescence intensities of the spots are shown in false-color, with white indicating saturated signal intensity, red indicating high signal intensity, green indicating moderate signal intensity, and blue indicating low signal intensity. (c) Zoom-in on a portion of the whole-genome yeast intergenic microarray bound by Rap1.
Figure 1. Protein binding microarray (PBM) schematic
(a) Overview of PBM experiments. (b) Whole-genome yeast intergenic microarray bound by Rap1. The fluorescence intensities of the spots are shown in false-color, with white indicating saturated signal intensity, red indicating high signal intensity, green indicating moderate signal intensity, and blue indicating low signal intensity. (c) Zoom-in on a portion of the whole-genome yeast intergenic microarray bound by Rap1.
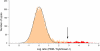
Figure 2. Identifying the specifically bound spots
(a) Distribution of Rap1 PBM ratio (PBM / SybrGreen I) data. The arrow indicates those spots passing a P value threshold of 0.001 after correction for multiple hypothesis testing. Indicated in red are spots harboring an exact match to a sequence belonging to our discovered Rap1 binding site motif. (b) Zoom-in on intergenic regions, from both PBMs (left) and SybrGreen I stained microarrays (right), upstream of RPL14A, RPL8A, and OPI3, which are known direct targets of Rap1. The fluorescence intensities of the spots are shown in false-color, color-coded as described previously (Fig. 1 legend). PBM P values are corrected for multiple hypotheses. Determination of binding in ChIP-chip experiments (“YES” or “NO”) is described in Methods. All regions shown have an exact match to a sequence belonging to the discovered Rap1 motif (“YES”). For each region, the binding site is conserved across five sensu stricto yeast strains either to within two standard deviations (“2 SD”) or 100% identical at each position (“Exact”) as described in Methods. “*” indicates Rap1 ChIP-chip data from Lee et al., and “#” indicates Rap1 ChIP-chip data from Lieb et al..
Figure 2. Identifying the specifically bound spots
(a) Distribution of Rap1 PBM ratio (PBM / SybrGreen I) data. The arrow indicates those spots passing a P value threshold of 0.001 after correction for multiple hypothesis testing. Indicated in red are spots harboring an exact match to a sequence belonging to our discovered Rap1 binding site motif. (b) Zoom-in on intergenic regions, from both PBMs (left) and SybrGreen I stained microarrays (right), upstream of RPL14A, RPL8A, and OPI3, which are known direct targets of Rap1. The fluorescence intensities of the spots are shown in false-color, color-coded as described previously (Fig. 1 legend). PBM P values are corrected for multiple hypotheses. Determination of binding in ChIP-chip experiments (“YES” or “NO”) is described in Methods. All regions shown have an exact match to a sequence belonging to the discovered Rap1 motif (“YES”). For each region, the binding site is conserved across five sensu stricto yeast strains either to within two standard deviations (“2 SD”) or 100% identical at each position (“Exact”) as described in Methods. “*” indicates Rap1 ChIP-chip data from Lee et al., and “#” indicates Rap1 ChIP-chip data from Lieb et al..
Figure 3. DNA binding site motifs as determined by PBMs compared to motifs derived from ChIP-chip data and from TRANSFAC
Sequence logos were generated essentially as described previously. Group specificity scores were calculated as described in Methods. “*” indicates Rap1, Abf1, and Mig1 ChIP-chip data from Lee et al., and “#” indicates Rap1 ChIP-chip data from Lieb et al.. Although the Mig1 binding site motif derived from the ChIP-chip data has a statistically significant group specificity score, it is not a match to either the TRANSFAC or PBM Mig1 motif. The Pearson correlation coefficients comparing the PBM versus ChIP-chip motifs, as well as those comparing each of these motifs versus the motifs present in the TRANSFAC database, were as follows: Rap1 PBM versus Lee et al. ChIP-chip: 0.992; Rap1 PBM versus Lieb et al. ChIP-chip: 0.995; Rap1 PBM versus TRANSFAC: 0.953; Rap1 Lee et al. versus Lieb et al. ChIP-chip: 0.985; Rap1 Lee et al. ChIP-chip versus TRANSFAC: 0.921; Rap1 Lieb et al. ChIP-chip versus TRANSFAC: 0.950; Abf1 PBM versus ChIP-chip: 0.989; Abf1 PBM versus TRANSFAC: 0.978; Abf1 ChIP-chip versus TRANSFAC: 0.986; Mig1 PBM versus ChIP-chip: 0.453; Mig1 PBM versus TRANSFAC: 0.938; Mig1 ChIP-chip versus TRANSFAC: 0.406.
Figure 4. EMSAs of PBM-derived Rap1 binding site sequences
(a) Rap1 binding site sequences present within the DNA probes corresponding to portions of the intergenic regions iYLL051C (P = 3.20 × 10-16) and iYPL221W (P = 3.91 x 10-21), aligned against the TRANSFAC and PBM-derived Rap1 binding site sequence logos. (b) Lanes 1 and 2, positive control DNA probe; lanes 3 and 4, negative control DNA probe; lanes 5 and 6, DNA probe corresponding to the best Rap1 binding site sequence that could be identified within the iYLL051C intergenic region; lanes 7 and 8, DNA probe corresponding to the PBM-derived Rap1 binding site sequence within the iYPL221W intergenic region. “-” indicates Rap1 protein was not present in the binding reaction; “+” indicates Rap1 protein was present the binding reaction.
Figure 4. EMSAs of PBM-derived Rap1 binding site sequences
(a) Rap1 binding site sequences present within the DNA probes corresponding to portions of the intergenic regions iYLL051C (P = 3.20 × 10-16) and iYPL221W (P = 3.91 x 10-21), aligned against the TRANSFAC and PBM-derived Rap1 binding site sequence logos. (b) Lanes 1 and 2, positive control DNA probe; lanes 3 and 4, negative control DNA probe; lanes 5 and 6, DNA probe corresponding to the best Rap1 binding site sequence that could be identified within the iYLL051C intergenic region; lanes 7 and 8, DNA probe corresponding to the PBM-derived Rap1 binding site sequence within the iYPL221W intergenic region. “-” indicates Rap1 protein was not present in the binding reaction; “+” indicates Rap1 protein was present the binding reaction.
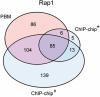
Figure 5. Comparison of bound intergenic regions derived from PBM data as compared to those derived from ChIP-chip,
Venn diagrams depicting the results of the comparison for (a) Rap1, (b) Abf1, and (c) Mig1. The Venn diagrams depict data only for those intergenic regions for which data were available for both ChIP-chip and PBMs. “*” indicates Rap1, Abf1, and Mig1 ChIP-chip data from Lee et al., and “#” indicates Rap1 ChIP-chip data from Lieb et al..
Figure 5. Comparison of bound intergenic regions derived from PBM data as compared to those derived from ChIP-chip,
Venn diagrams depicting the results of the comparison for (a) Rap1, (b) Abf1, and (c) Mig1. The Venn diagrams depict data only for those intergenic regions for which data were available for both ChIP-chip and PBMs. “*” indicates Rap1, Abf1, and Mig1 ChIP-chip data from Lee et al., and “#” indicates Rap1 ChIP-chip data from Lieb et al..
Figure 5. Comparison of bound intergenic regions derived from PBM data as compared to those derived from ChIP-chip,
Venn diagrams depicting the results of the comparison for (a) Rap1, (b) Abf1, and (c) Mig1. The Venn diagrams depict data only for those intergenic regions for which data were available for both ChIP-chip and PBMs. “*” indicates Rap1, Abf1, and Mig1 ChIP-chip data from Lee et al., and “#” indicates Rap1 ChIP-chip data from Lieb et al..
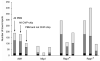
Figure 6. Cross-species sequence conservation of binding sites identified from PBM data as compared to those identified from ChIP-chip data
From left to right for a single TF, bars represent all spots bound in PBMs, all spots bound in ChIP-chip, and spots bound in PBMs and not ChIP-chip. Spots were called `bound' as described in Methods. Shown in dark gray is the subset of bound spots with S. cerevisiae binding sites conserved to within two standard deviations of the motif average across all five sensu stricto species. Shown in black is the subset of S. cerevisiae bound spots with conserved sites 100% identical across all five species. The remaining bound spots are in light gray.
Similar articles
- Protein binding microarrays (PBMs) for rapid, high-throughput characterization of the sequence specificities of DNA binding proteins.
Berger MF, Bulyk ML. Berger MF, et al. Methods Mol Biol. 2006;338:245-60. doi: 10.1385/1-59745-097-9:245. Methods Mol Biol. 2006. PMID: 16888363 Free PMC article. - Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors.
Berger MF, Bulyk ML. Berger MF, et al. Nat Protoc. 2009;4(3):393-411. doi: 10.1038/nprot.2008.195. Nat Protoc. 2009. PMID: 19265799 Free PMC article. - Protein binding microarrays for the characterization of DNA-protein interactions.
Bulyk ML. Bulyk ML. Adv Biochem Eng Biotechnol. 2007;104:65-85. doi: 10.1007/10_025. Adv Biochem Eng Biotechnol. 2007. PMID: 17290819 Free PMC article. Review. - Analysis of sequence specificities of DNA-binding proteins with protein binding microarrays.
Bulyk ML. Bulyk ML. Methods Enzymol. 2006;410:279-99. doi: 10.1016/S0076-6879(06)10013-0. Methods Enzymol. 2006. PMID: 16938556 Free PMC article. Review. - Linking DNA-binding proteins to their recognition sequences by using protein microarrays.
Ho SW, Jona G, Chen CT, Johnston M, Snyder M. Ho SW, et al. Proc Natl Acad Sci U S A. 2006 Jun 27;103(26):9940-5. doi: 10.1073/pnas.0509185103. Epub 2006 Jun 19. Proc Natl Acad Sci U S A. 2006. PMID: 16785442 Free PMC article.
Cited by
- STAT1:DNA sequence-dependent binding modulation by phosphorylation, protein:protein interactions and small-molecule inhibition.
Bonham AJ, Wenta N, Osslund LM, Prussin AJ 2nd, Vinkemeier U, Reich NO. Bonham AJ, et al. Nucleic Acids Res. 2013 Jan;41(2):754-63. doi: 10.1093/nar/gks1085. Epub 2012 Nov 24. Nucleic Acids Res. 2013. PMID: 23180800 Free PMC article. - Improved models for transcription factor binding site identification using nonindependent interactions.
Zhao Y, Ruan S, Pandey M, Stormo GD. Zhao Y, et al. Genetics. 2012 Jul;191(3):781-90. doi: 10.1534/genetics.112.138685. Epub 2012 Apr 13. Genetics. 2012. PMID: 22505627 Free PMC article. - Emergence of the Noncoding Cancer Genome: A Target of Genetic and Epigenetic Alterations.
Zhou S, Treloar AE, Lupien M. Zhou S, et al. Cancer Discov. 2016 Nov;6(11):1215-1229. doi: 10.1158/2159-8290.CD-16-0745. Epub 2016 Oct 19. Cancer Discov. 2016. PMID: 27807102 Free PMC article. Review. - Evidence-ranked motif identification.
Georgiev S, Boyle AP, Jayasurya K, Ding X, Mukherjee S, Ohler U. Georgiev S, et al. Genome Biol. 2010;11(2):R19. doi: 10.1186/gb-2010-11-2-r19. Epub 2010 Feb 15. Genome Biol. 2010. PMID: 20156354 Free PMC article. - Connecting protein structure with predictions of regulatory sites.
Morozov AV, Siggia ED. Morozov AV, et al. Proc Natl Acad Sci U S A. 2007 Apr 24;104(17):7068-73. doi: 10.1073/pnas.0701356104. Epub 2007 Apr 16. Proc Natl Acad Sci U S A. 2007. PMID: 17438293 Free PMC article.
References
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. - PubMed
- Wodicka L, Dong H, Mittmann M, Ho MH, Lockhart DJ. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 1997;15:1359–1367. - PubMed
- Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. - PubMed
- Iyer VR, et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. - PubMed
- Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 2001;28:327–334. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous