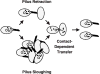Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems - PubMed (original) (raw)
Review
Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems
Peter J Christie. Biochim Biophys Acta. 2004.
Abstract
The translocation of DNA across biological membranes is an essential process for many living organisms. In bacteria, type IV secretion systems (T4SS) are used to deliver DNA as well as protein substrates from donor to target cells. The T4SS are structurally complex machines assembled from a dozen or more membrane proteins in response to environmental signals. In Gram-negative bacteria, the conjugation machines are composed of a cell envelope-spanning secretion channel and an extracellular pilus. These dynamic structures (i) direct formation of stable contacts-the mating junction-between donor and recipient cell membranes, (ii) transmit single-stranded DNA as a nucleoprotein particle, as well as protein substrates, across donor and recipient cell membranes, and (iii) mediate disassembly of the mating junction following substrate transfer. This review summarizes recent progress in our understanding of the mechanistic details of DNA trafficking with a focus on the paradigmatic Agrobacterium tumefaciens VirB/D4 T4SS and related conjugation systems.
Figures
Fig. 1
T4SS conjugally transfer DNA and protein substrates to target cells, as illustrated for the A. tumefaciens VirB/D4 T4SS. The transfer DNA (T-DNA) is processed by Dtr proteins (VirD1, VirD2, and VirC1) bound at _oriT_-like border repeat sequences to form the VirD2-T-strand transfer intermediate. The VirB/D4 T4SS recruits the DNA transfer intermediate through binding of the VirD4 coupling protein (CP) to the positively charged C terminus of VirD2, which is unfolded prior to export. The transfer intermediate is translocated across the cell envelope through a secretion channel composed of the VirD4 CP and the VirB mating pair formation (Mpf) proteins; the precise route of translocation is undefined. Translocation of protein substrates similarly proceeds in three steps: (i) processing, which is independent or dependent on chaperone binding to maintain substrate in an unfolded, nonaggregated conformation; (ii) recruitment, mediated by CP binding to positively charged C-termini; and (iii) translocation, via the CP/Mpf secretion channel. IM, inner membrane; P, periplasm; OM, outer membrane.
Fig. 2
Type IV secretion substrates contain clusters of positively charged residues at their C termini. This charge bias is a postulated feature of the type IV secretion signal recognized by the coupling protein component of the T4SS [31,33,35]. Shown are the C-terminal residues, with Arg (R), Lys (K), and His (H) residues highlighted with box shading and the Arg-X-Arg motifs of VirB/D4 T4SS substrates with wavy underlines. Known or potential substrates include VirB/D4 T4SS substrates encoded by the pTiC58 (C58) and pTiA6NC (A6) plasmids of A. tumefaciens, relaxases encoded by the plasmids listed, CagA exported by the H. pylori Cag T4SS, and RalF exported by the L. pneumophila Dot/Icm T4SS. VirD3 proteins from the C58 and A6 plasmids are candidate VirB/D4 secretion substrates on the basis of a C-terminal charge bias. VirE1 is not predicted to be a substrate on the basis of charge distribution, in agreement with findings that this secretion chaperone is not recruited by the VirD4 CP to the VirB/D4 T4SS [32,35].
Fig. 3
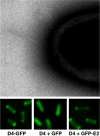
The A. tumefaciens T4SS and a secretion substrate localize at the cell poles. The T pili localize at cell poles as detected by electron microscopy (kindly provided by Alain Bernadac), the VirD4 CP localizes at the cell poles as shown by polar fluorescence of a VirD4-GFP fusion protein, and the VirD4 CP recruits VirE2 to the cell poles as shown by VirD4-dependent polar fluorescence of GFP-VirE2. GFP itself distributes throughout the cytoplasm of A. tumefaciens cells (middle panel) (see Ref. [35]).
Fig. 4
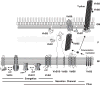
Topologies and oligomeric structures of VirB/D4 T4SS components. The VirD4 CP undergoes monomer–hexamer transitions possibly mediated by ATP binding; the VirB11 ATPase undergoes dramatic conformational changes in its N-terminal domain as a function of ATP binding and ADP release; VirB4 is also postulated to bind and hydrolyze ATP. These components plus the channel subunits shown are postulated to form a membrane-spanning secretion channel. A disulfide-linked dimer of VirB7 lipoprotein and secretin-like VirB9 are postulated to form a pore at the OM for pilus protusion and/or substrate transfer. Independently of the VirD4 CP, the VirB proteins direct polymerization of the T pilus, which may or may not be physically joined to the secretion channel. IM, inner membrane; P, periplasm; OM, outer membrane.
Fig. 5
A model depicting the A. tumefaciens VirB/D4 T4SS as a single, supramolecular organelle. The VirD4 CP is a homomultimeric integral membrane complex required for substrate transfer. The VirB proteins assemble as a secretion channel and an extracellular T pilus. VirD4 and the VirB proteins function together to mediate substrate transfer, and the VirB proteins direct pilus assembly. Cells are postulated to shed the adhesive T pilus as a means of promoting aggregation of donor and recipients on solid surfaces. IM, inner membrane; P, periplasm; OM, outer membrane.
Fig. 6
Conjugative pili likely function by a retraction mechanism, e.g., F pilus, or a sloughing mechanism, e.g., RP4, R388, VirB/D4 pili. Transfer systems utilizing the latter mechanism shed their pili presumably to induce aggregation of donor and recipient cells. Both mechanisms bring donor and recipient cell membranes into close apposition for formation of mating junctions. Contact between donor and recipient membranes stimulates channel opening and substrate transfer. Following transfer, the mating junction disassembles by an unknown mechanism(s). “Uncoupling” mutations of the VirB/D4 T4SS permit substrate transfer on solid surfaces in the absence of detectable pilus production [80,81].
Similar articles
- Structural and dynamic properties of bacterial type IV secretion systems (review).
Christie PJ, Cascales E. Christie PJ, et al. Mol Membr Biol. 2005 Jan-Apr;22(1-2):51-61. doi: 10.1080/09687860500063316. Mol Membr Biol. 2005. PMID: 16092524 Free PMC article. Review. - The Agrobacterium VirB/VirD4 T4SS: Mechanism and Architecture Defined Through In Vivo Mutagenesis and Chimeric Systems.
Li YG, Christie PJ. Li YG, et al. Curr Top Microbiol Immunol. 2018;418:233-260. doi: 10.1007/82_2018_94. Curr Top Microbiol Immunol. 2018. PMID: 29808338 Free PMC article. Review. - Promiscuous DNA transfer system of Agrobacterium tumefaciens: role of the virB operon in sex pilus assembly and synthesis.
Kado CI. Kado CI. Mol Microbiol. 1994 Apr;12(1):17-22. doi: 10.1111/j.1365-2958.1994.tb00990.x. Mol Microbiol. 1994. PMID: 7914664 Review. - Definition of a bacterial type IV secretion pathway for a DNA substrate.
Cascales E, Christie PJ. Cascales E, et al. Science. 2004 May 21;304(5674):1170-3. doi: 10.1126/science.1095211. Science. 2004. PMID: 15155952 Free PMC article. - The mating pair formation system of conjugative plasmids-A versatile secretion machinery for transfer of proteins and DNA.
Schröder G, Lanka E. Schröder G, et al. Plasmid. 2005 Jul;54(1):1-25. doi: 10.1016/j.plasmid.2005.02.001. Plasmid. 2005. PMID: 15907535 Review.
Cited by
- Evidence for VirB4-mediated dislocation of membrane-integrated VirB2 pilin during biogenesis of the Agrobacterium VirB/VirD4 type IV secretion system.
Kerr JE, Christie PJ. Kerr JE, et al. J Bacteriol. 2010 Oct;192(19):4923-34. doi: 10.1128/JB.00557-10. Epub 2010 Jul 23. J Bacteriol. 2010. PMID: 20656905 Free PMC article. - The Love and Hate Relationship between T5SS and Other Secretion Systems in Bacteria.
Luo Y, Chen Z, Lian S, Ji X, Zhu C, Zhu G, Xia P. Luo Y, et al. Int J Mol Sci. 2023 Dec 24;25(1):281. doi: 10.3390/ijms25010281. Int J Mol Sci. 2023. PMID: 38203452 Free PMC article. - Global transcriptome analysis of Mesorhizobium alhagi CCNWXJ12-2 under salt stress.
Liu X, Luo Y, Mohamed OA, Liu D, Wei G. Liu X, et al. BMC Microbiol. 2014 Dec 24;14:1. doi: 10.1186/s12866-014-0319-y. BMC Microbiol. 2014. PMID: 25539655 Free PMC article. - Mechanism and Function of Type IV Secretion During Infection of the Human Host.
Gonzalez-Rivera C, Bhatty M, Christie PJ. Gonzalez-Rivera C, et al. Microbiol Spectr. 2016 Jun;4(3):10.1128/microbiolspec.VMBF-0024-2015. doi: 10.1128/microbiolspec.VMBF-0024-2015. Microbiol Spectr. 2016. PMID: 27337453 Free PMC article. Review. - Conjugative Mating Assays for Sequence-specific Analysis of Transfer Proteins Involved in Bacterial Conjugation.
Erdogan F, Lento C, Yaseen A, Nowroozi-Dayeni R, Kheyson S, Audette GF. Erdogan F, et al. J Vis Exp. 2017 Jan 4;(119):54854. doi: 10.3791/54854. J Vis Exp. 2017. PMID: 28117821 Free PMC article.
References
- Lawley TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. - PubMed
- Dillard JP, Seifert HS. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol. 2001;41:263–277. - PubMed
- Hofreuter D, Odenbreit S, Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol. 2001;41:379–391. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources