The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle - PubMed (original) (raw)
The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle
Yael Aylon et al. EMBO J. 2004.
Abstract
DNA double-strand breaks (DSBs) are dangerous lesions that can lead to genomic instability and cell death. Eukaryotic cells repair DSBs either by nonhomologous end-joining (NHEJ) or by homologous recombination. We investigated the ability of yeast cells (Saccharomyces cerevisiae) to repair a single, chromosomal DSB by recombination at different stages of the cell cycle. We show that cells arrested at the G1 phase of the cell cycle restrict homologous recombination, but are able to repair the DSB by NHEJ. Furthermore, we demonstrate that recombination ability does not require duplicated chromatids or passage through S phase, and is controlled at the resection step by Clb-CDK activity.
Figures
Figure 1
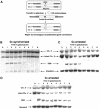
G1-arrested cells are unable to repair a DSB. (A) Schematic representation of our experimental system. Open rectangles represent the ura3 alleles on chromosomes II and V. A black box represents the HOcs; a grey box depicts the inactive HOcs-inc flanked by the _Bam_HI (designated B) and _Eco_RI (designated R) restriction sites. Transfer of the cells to galactose-containing medium results in a DSB that is repaired by homologous recombination. (B) Southern blot analysis of cells synchronized in G1 with alpha-factor and released from the arrest at _t_=0 in medium containing galactose. The genomic DNA was digested with _Cla_I and probed with a 1.2 kb fragment containing the URA3 gene (open rectangles in (A)). The donor sequences on chromosome II, the intact chromosome V and one of the two bands created by the DSB are shown. A probe complementary to LEU2 sequences on chromosome III served as a loading standard. (C) Southern blot analysis of DNA from cells arrested at G2 with nocodazole and transferred to galactose-containing medium in the presence of nocodazole. (D) Southern blot analysis of DNA from cells arrested at G1 with alpha-factor and transferred to galactose-containing medium in the presence of alpha-factor.
Figure 2
G1-arrested cells do not carry out gene conversion and cells synchronized at G1 delay gene conversion. (A) PCR-based assay for the detection of gene conversion. The region flanking the site of the DSB was amplified by PCR. The products were digested with _Bam_HI and subjected to electrophoresis. The presence of a _Bam_HI site demonstrates repair by gene conversion. (B) Kinetics of gene conversion of MK203 cells at different stages of the cell cycle. The ratio between _Bam_HI-cut and total PCR products was plotted as a function of time.
Figure 3
DSB ends remain unresected in G1-arrested cells and are repaired by NHEJ. (A) G1-synchronized and G1-arrested cells were incubated in galactose-containing medium for 6 h and plated on glucose-containing medium. The fate of the broken chromosomes in randomly picked individual colonies was analysed by PCR as described above. (B) Nondenaturing slot blot analysis was carried out using a 1.2 kb fragment of the URA3 gene (open rectangle in Figure 1A). Resected DNA hybridizes to the probe. (C) Southern blot analysis of genomic DNA from G1-arrested dnl4 cells in galactose.
Figure 4
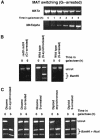
G1-arrested cells are able to carry out MAT switching; the inability to repair DSBs by homologous recombination in G1 is independent of checkpoint response and ploidy. (A) A MATa derivative of MK203 was retained at G1 (alpha-factor) and monitored for the ability to switch mating type with _MATalpha-_specific PCR primers. (B) PCR assays for gene conversion were carried out for rad9 rad24 synchronized or arrested in G1, and wt cells synchronized or arrested in S-phase with HU. (C) Disomic and diploid strains are unable to carry out gene conversion when arrested at G1 (alpha-factor) or S (HU). PCR was carried out as in (B) and PCR products were digested with _Bam_HI and _Nco_I. The presence of uncut product at _t_=6 h indicates that no gene conversion occurred.
Figure 5
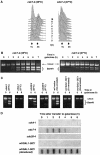
Homologous recombination is dependent on active Clb–Cdk and not on DNA replication. (A) cdc7-4 cells were arrested at the restrictive temperature and then incubated in galactose-containing medium at the permissive (25°C) or restrictive (37°C) temperatures. FACS analysis of the cdc7-4 cell culture confirmed that cells did not undergo DNA replication at the restrictive temperature. (B) PCR assays for gene conversion in cdc7-4 cells at the permissive and restrictive temperatures. (C) PCR assays for gene conversion were carried out for cdc4-1 and cdc28-4 cells at the permissive and restrictive temperatures, wt G2-arrested with vector only, or G1- or G2-arrested with GAL-SIC1 plasmid. (D) Nondenaturing slot blot analysis of cdc4-1, cdc7-4, cdc28-4 and wt cells overexpressing SIC1. As a positive control for hybridization, equivalent DNA samples were blotted after DNA denaturation (only wt/GAL-SIC is shown here).
Similar articles
- Differential usage of non-homologous end-joining and homologous recombination in double strand break repair.
Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Sonoda E, et al. DNA Repair (Amst). 2006 Sep 8;5(9-10):1021-9. doi: 10.1016/j.dnarep.2006.05.022. Epub 2006 Jun 27. DNA Repair (Amst). 2006. PMID: 16807135 Review. - Cyclin-dependent kinase-dependent phosphorylation of Lif1 and Sae2 controls imprecise nonhomologous end joining accompanied by double-strand break resection.
Matsuzaki K, Terasawa M, Iwasaki D, Higashide M, Shinohara M. Matsuzaki K, et al. Genes Cells. 2012 Jun;17(6):473-93. doi: 10.1111/j.1365-2443.2012.01602.x. Epub 2012 May 8. Genes Cells. 2012. PMID: 22563681 - Non-homologous end-joining factors of Saccharomyces cerevisiae.
Dudásová Z, Dudás A, Chovanec M. Dudásová Z, et al. FEMS Microbiol Rev. 2004 Nov;28(5):581-601. doi: 10.1016/j.femsre.2004.06.001. FEMS Microbiol Rev. 2004. PMID: 15539075 Review. - Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells.
Saleh-Gohari N, Helleday T. Saleh-Gohari N, et al. Nucleic Acids Res. 2004 Jul 13;32(12):3683-8. doi: 10.1093/nar/gkh703. Print 2004. Nucleic Acids Res. 2004. PMID: 15252152 Free PMC article.
Cited by
- A high-throughput scintillation proximity-based assay for human DNA ligase IV.
Tseng HM, Shum D, Bhinder B, Escobar S, Veomett NJ, Tomkinson AE, Gin DY, Djaballah H, Scheinberg DA. Tseng HM, et al. Assay Drug Dev Technol. 2012 Jun;10(3):235-49. doi: 10.1089/adt.2011.0404. Epub 2011 Dec 22. Assay Drug Dev Technol. 2012. PMID: 22192310 Free PMC article. - DNA Repair Pathway Choices in CRISPR-Cas9-Mediated Genome Editing.
Xue C, Greene EC. Xue C, et al. Trends Genet. 2021 Jul;37(7):639-656. doi: 10.1016/j.tig.2021.02.008. Epub 2021 Apr 22. Trends Genet. 2021. PMID: 33896583 Free PMC article. Review. - Mating-type genes and MAT switching in Saccharomyces cerevisiae.
Haber JE. Haber JE. Genetics. 2012 May;191(1):33-64. doi: 10.1534/genetics.111.134577. Genetics. 2012. PMID: 22555442 Free PMC article. Review. - ATR-mediated proteome remodeling is a major determinant of homologous recombination capacity in cancer cells.
Kim D, Liu Y, Oberly S, Freire R, Smolka MB. Kim D, et al. Nucleic Acids Res. 2018 Sep 19;46(16):8311-8325. doi: 10.1093/nar/gky625. Nucleic Acids Res. 2018. PMID: 30010936 Free PMC article. - DNA damage triggers increased mobility of chromosomes in G1-phase cells.
Smith MJ, Bryant EE, Joseph FJ, Rothstein R. Smith MJ, et al. Mol Biol Cell. 2019 Oct 1;30(21):2620-2625. doi: 10.1091/mbc.E19-08-0469. Epub 2019 Sep 4. Mol Biol Cell. 2019. PMID: 31483739 Free PMC article.
References
- Bartek J, Lukas J, Bartkova J (1999) Perspective: defects in cell cycle control and cancer. J Pathol 187: 95–99 - PubMed
- Fabre F, Boulet A, Roman H (1984) Gene conversion at different points in the mitotic cycle of Saccharomyces cerevisiae. Mol Gen Genet 195: 139–143 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases