Long-range control of gene expression: emerging mechanisms and disruption in disease - PubMed (original) (raw)
Review
Long-range control of gene expression: emerging mechanisms and disruption in disease
Dirk A Kleinjan et al. Am J Hum Genet. 2005 Jan.
Abstract
Transcriptional control is a major mechanism for regulating gene expression. The complex machinery required to effect this control is still emerging from functional and evolutionary analysis of genomic architecture. In addition to the promoter, many other regulatory elements are required for spatiotemporally and quantitatively correct gene expression. Enhancer and repressor elements may reside in introns or up- and downstream of the transcription unit. For some genes with highly complex expression patterns--often those that function as key developmental control genes--the cis-regulatory domain can extend long distances outside the transcription unit. Some of the earliest hints of this came from disease-associated chromosomal breaks positioned well outside the relevant gene. With the availability of wide-ranging genome sequence comparisons, strong conservation of many noncoding regions became obvious. Functional studies have shown many of these conserved sites to be transcriptional regulatory elements that sometimes reside inside unrelated neighboring genes. Such sequence-conserved elements generally harbor sites for tissue-specific DNA-binding proteins. Developmentally variable chromatin conformation can control protein access to these sites and can regulate transcription. Disruption of these finely tuned mechanisms can cause disease. Some regulatory element mutations will be associated with phenotypes distinct from any identified for coding-region mutations.
Figures
Figure 1
Schematic representation of a theoretical gene locus, highlighting various cis elements that contribute to the regulation of gene expression. Exons are indicated by rectangular boxes, with the protein-coding portions in black. Complexity of gene output can be achieved through use of alternative promoters and/or exons. Multiple _cis_-regulatory elements, indicated by ovals, control the quantitative and spatiotemporal specific expression. These elements may be at considerable distances from the promoter, either upstream or downstream, and are sometimes within or beyond an adjacent gene. The chromatin structure of the locus is determined by a combination of the activities of these cis elements and the wider chromosomal and nuclear environment. In some loci, the outermost cis elements may carry some boundary activity, isolating the specific chromatin structure of the gene domain from that of adjacent chromosomal segments.
Figure 2
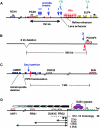
Details of position-effect cases caused by disruption of long-range gene control. In all cases, the affected gene(s) are shown in red, and other genes are shown in purple or blue. Filled boxes indicate individual exons, and hashed boxes represent full genes. L-shaped black arrows indicate the direction of transcription. A, Human PAX6 locus. The loss of a set of DNase I HSs downstream from one allele causes aniridia. The HSs are located within introns of the adjacent ubiquitously expressed ELP4 gene. Some documented aniridia-associated breakpoints are denoted by blue arrows. The downstream end of the correcting YAC transgene (YA) and the noncorrecting one (YB) are shown in green. Both upstream YAC ends are ∼200 kb 5′ of the PAX6 promoters. Isolated HSs have been shown to act as tissue-specific enhancers for lens and retinal expression. B, The human POU3F4 deafness locus. The microdeletion of an 8-kb region located 900 kb upstream of the gene contains a conserved noncoding sequence, the loss of which leads to congenital deafness. The mouse slf inversion breakpoint X leaves the neural tube enhancer (nt) intact. C, Mouse/human upstream SHH region. A complex hotspot for limb abnormalities is found 1 Mb upstream of SHH, within the introns of LMBR1. The region contains a conserved noncoding element that is capable of functioning as an enhancer that drives SHH expression in the limb bud in both an anterior and posterior zone, as well as a repressor element that silences the anterior expression. The Sasquatch insertion disrupts the anterior repression function, whereas the acheiropodia deletion is thought to disrupt positive enhancer activity. D, Human FSHD region. Deletion of an integral number of D4Z4 repeats from the tip of the long arm of chromosome 4 to below a threshold of 10 repeats results in FSHD. A contentious model suggests that a multiprotein repressor complex fails to bind adequately to the deleted allele, which leads to derepression of several genes in the region proximal to the repeat array and causes the phenotype. E, Human α-globin locus (HBA). Deletion of the polyadenylation signal from the ubiquitously expressed LUC7L gene on the opposite strand leads to transcription of an antisense RNA that runs through the HBA2 gene, resulting in silencing and methylation of the HBA2 promoter. Open ovals indicate unmethylated CpG islands; the gray oval depicts the methylated CpG island. F, Mouse Hoxd cluster. A GCR regulates expression of multiple consecutive Hoxd genes in a tissue-specific manner. In the distal limb, the GCR also regulates the expression of Lnp, Evx2, and Hoxd13–10, whereas in the CNS it controls Lnp and Evx2. G, Mouse IL4/IL13 region. A conserved noncoding element (CNE) located between IL4 and IL13 controls expression of both genes, as well as IL5, but does not influence expression of the KIF3a and RAD50 genes. H, Human β-globin locus (HBB). Deletion of a large genomic region upstream of the human β-globin genes, including the LCR, results in reduced DNase I sensitivity and histone acetylation levels across the locus, which causes loss of globin expression. The β-globin locus is embedded within a region that contains numerous OR genes.
Figure 2
Details of position-effect cases caused by disruption of long-range gene control. In all cases, the affected gene(s) are shown in red, and other genes are shown in purple or blue. Filled boxes indicate individual exons, and hashed boxes represent full genes. L-shaped black arrows indicate the direction of transcription. A, Human PAX6 locus. The loss of a set of DNase I HSs downstream from one allele causes aniridia. The HSs are located within introns of the adjacent ubiquitously expressed ELP4 gene. Some documented aniridia-associated breakpoints are denoted by blue arrows. The downstream end of the correcting YAC transgene (YA) and the noncorrecting one (YB) are shown in green. Both upstream YAC ends are ∼200 kb 5′ of the PAX6 promoters. Isolated HSs have been shown to act as tissue-specific enhancers for lens and retinal expression. B, The human POU3F4 deafness locus. The microdeletion of an 8-kb region located 900 kb upstream of the gene contains a conserved noncoding sequence, the loss of which leads to congenital deafness. The mouse slf inversion breakpoint X leaves the neural tube enhancer (nt) intact. C, Mouse/human upstream SHH region. A complex hotspot for limb abnormalities is found 1 Mb upstream of SHH, within the introns of LMBR1. The region contains a conserved noncoding element that is capable of functioning as an enhancer that drives SHH expression in the limb bud in both an anterior and posterior zone, as well as a repressor element that silences the anterior expression. The Sasquatch insertion disrupts the anterior repression function, whereas the acheiropodia deletion is thought to disrupt positive enhancer activity. D, Human FSHD region. Deletion of an integral number of D4Z4 repeats from the tip of the long arm of chromosome 4 to below a threshold of 10 repeats results in FSHD. A contentious model suggests that a multiprotein repressor complex fails to bind adequately to the deleted allele, which leads to derepression of several genes in the region proximal to the repeat array and causes the phenotype. E, Human α-globin locus (HBA). Deletion of the polyadenylation signal from the ubiquitously expressed LUC7L gene on the opposite strand leads to transcription of an antisense RNA that runs through the HBA2 gene, resulting in silencing and methylation of the HBA2 promoter. Open ovals indicate unmethylated CpG islands; the gray oval depicts the methylated CpG island. F, Mouse Hoxd cluster. A GCR regulates expression of multiple consecutive Hoxd genes in a tissue-specific manner. In the distal limb, the GCR also regulates the expression of Lnp, Evx2, and Hoxd13–10, whereas in the CNS it controls Lnp and Evx2. G, Mouse IL4/IL13 region. A conserved noncoding element (CNE) located between IL4 and IL13 controls expression of both genes, as well as IL5, but does not influence expression of the KIF3a and RAD50 genes. H, Human β-globin locus (HBB). Deletion of a large genomic region upstream of the human β-globin genes, including the LCR, results in reduced DNase I sensitivity and histone acetylation levels across the locus, which causes loss of globin expression. The β-globin locus is embedded within a region that contains numerous OR genes.
Figure 3
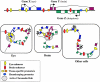
Model for the coexistence of physically overlapping but independently regulated “functional gene expression modules” in the same genomic region. A hypothetical region containing two tissue-specific genes and one housekeeping gene. Gene X (blue exons) is expressed in eye tissue, gene Y (purple exons) is expressed in brain, and gene Z (green exons) is ubiquitously expressed. Transcriptional activity depends on the formation of an ACH that encompasses tissue-specific _cis_-acting elements with bound transcription factor complexes and selective interaction with the relevant gene promoter. Formation of an ACH provides a high local concentration of transcription factors and positive chromatin-modifying enzymes. The housekeeping promoter is active in all cells and does not rely on tissue-specific ACH formation.
Similar articles
- Transcriptional rewiring of the sex determining dmrt1 gene duplicate by transposable elements.
Herpin A, Braasch I, Kraeussling M, Schmidt C, Thoma EC, Nakamura S, Tanaka M, Schartl M. Herpin A, et al. PLoS Genet. 2010 Feb 12;6(2):e1000844. doi: 10.1371/journal.pgen.1000844. PLoS Genet. 2010. PMID: 20169179 Free PMC article. - Disruption of long-range gene regulation in human genetic disease: a kaleidoscope of general principles, diverse mechanisms and unique phenotypic consequences.
Bhatia S, Kleinjan DA. Bhatia S, et al. Hum Genet. 2014 Jul;133(7):815-45. doi: 10.1007/s00439-014-1424-6. Epub 2014 Feb 5. Hum Genet. 2014. PMID: 24496500 Review. - The developmental control of transposable elements and the evolution of higher species.
Friedli M, Trono D. Friedli M, et al. Annu Rev Cell Dev Biol. 2015;31:429-51. doi: 10.1146/annurev-cellbio-100814-125514. Epub 2015 Sep 17. Annu Rev Cell Dev Biol. 2015. PMID: 26393776 Review.
Cited by
- scPair: Boosting single cell multimodal analysis by leveraging implicit feature selection and single cell atlases.
Hu H, Quon G. Hu H, et al. Nat Commun. 2024 Nov 15;15(1):9932. doi: 10.1038/s41467-024-53971-2. Nat Commun. 2024. PMID: 39548084 Free PMC article. - Genotype inference from aggregated chromatin accessibility data reveals genetic regulatory mechanisms.
Wenz BM, He Y, Chen NC, Pickrell JK, Li JH, Dudek MF, Li T, Keener R, Voight BF, Brown CD, Battle A. Wenz BM, et al. bioRxiv [Preprint]. 2024 Sep 5:2024.09.04.610850. doi: 10.1101/2024.09.04.610850. bioRxiv. 2024. PMID: 39282458 Free PMC article. Preprint. - A hybrid approach for predicting transcription factors.
Patiyal S, Tiwari P, Ghai M, Dhapola A, Dhall A, Raghava GPS. Patiyal S, et al. Front Bioinform. 2024 Jul 25;4:1425419. doi: 10.3389/fbinf.2024.1425419. eCollection 2024. Front Bioinform. 2024. PMID: 39119181 Free PMC article. - The Effects of De Novo Mutation on Gene Expression and the Consequences for Fitness in Chlamydomonas reinhardtii.
Balogun EJ, Ness RW. Balogun EJ, et al. Mol Biol Evol. 2024 Mar 1;41(3):msae035. doi: 10.1093/molbev/msae035. Mol Biol Evol. 2024. PMID: 38366781 Free PMC article. - Enhancer mutations modulate the severity of chemotherapy-induced myelosuppression.
Zhigulev A, Norberg Z, Cordier J, Spalinskas R, Bassereh H, Björn N, Pradhananga S, Gréen H, Sahlén P. Zhigulev A, et al. Life Sci Alliance. 2024 Jan 16;7(3):e202302244. doi: 10.26508/lsa.202302244. Print 2024 Mar. Life Sci Alliance. 2024. PMID: 38228368 Free PMC article.
References
Electronic-Database Information
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for aniridia, deafness type 3, Rieger syndrome type 1, Greig cephalopolysyndactyly syndrome, MAF, LD, blepharophimosis-ptosis-epicanthus inversus syndrome, CD, HPE2, HPE3, PPD, SHFM1, FSHD, α-thalassemia, CDLS, and lactase expression)
References
- Bakker E, Wijmenga C, Vossen RH, Padberg GW, Hewitt J, van der WM, Rasmussen K, Frants RR (1995) The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve 2:S39–S44 - PubMed
- Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, Mitchell HF, Donis-Keller H, Helms C, Hing AV, Heng HH, Koop B, Martindale D, Rommens JM, Tsui LC, Scherer SW (1996) Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat Genet 14:353–35610.1038/ng1196-353 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources