Replication-independent chromatin loading of Dnmt1 during G2 and M phases - PubMed (original) (raw)
Replication-independent chromatin loading of Dnmt1 during G2 and M phases
Hariharan P Easwaran et al. EMBO Rep. 2004 Dec.
Abstract
The major DNA methyltransferase, Dnmt1, associates with DNA replication sites in S phase maintaining the methylation pattern in the newly synthesized strand. In view of the slow kinetics of Dnmt1 in vitro versus the fast progression of the replication fork, we have tested whether Dnmt1 associates with chromatin beyond S phase. Using time-lapse microscopy of mammalian cells expressing green-fluorescent-protein-tagged Dnmt1 and DsRed-tagged DNA Ligase I as a cell cycle progression marker, we have found that Dnmt1 associates with chromatin during G2 and M. This association is mediated by a specific targeting sequence, shows strong preference for constitutive but not facultative heterochromatin and is independent of heterochromatin-specific histone H3 Lys 9 trimethylation, SUV39H and HP1. Moreover, photobleaching analyses showed that Dnmt1 is continuously loaded onto chromatin throughout G2 and M, indicating a replication-independent role of Dnmt1 that could represent a novel and separate pathway to maintain DNA methylation.
Figures
Figure 1
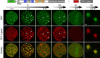
Dnmt1 shows cell-cycle-dependent association with chromatin. Live-cell imaging of C2C12 cells expressing full-length Dnmt1 fusion to GFP (GMT) and RFP–ligase. Selected frames from this time series are shown. At the time imaging was started (0 h), this cell was in mids phase. The arrowheads indicate preferential association of GMT with heterochromatic regions during late S and G2. (The images from mitosis and G1 in this assembly are from a different cell.) Scale bar, 5 μm.
Figure 2
Mapping of Dnmt1 domains responsible for replication foci association. DMAP corresponds to the DMAP1 transcriptional repressor interacting domain (Rountree et al, 2000). For other domains, see text. *Association takes place not only in S phase (see Fig 3).
Figure 3
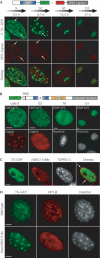
TS mediates association of Dnmt1 with chromatin, preferentially constitutive heterochromatin, which is independent of H3K9 trimethylation. (A) Live-cell imaging of C2C12 cells expressing F-TS–GFP and RFP–ligase. Selected frames from this time series are shown. After 3.5 h from start of imaging, the cell has entered late S phase (deciphered from the pattern of RFP–ligase) and the large F-TS–GFP foci (arrows) show colocalization with RFP–ligase. (B) C2C12 cells expressing GFP-tagged Dnmt1 lacking TS (GMTΔTS) analysed at the different cell cycle stages (for strategy, see supplementary Fig 1 online. (C) TS preferentially associates with constitutive heterochromatin and not with facultative heterochromatin. C2C12 cell expressing TS–GFP was immunostained with anti-trimethylated H3K27 antibody labelling the inactive X chromosome and DNA was visualized with TOPRO-3. These C2C12 cells are polyploid and have two inactive X chromosomes (H.P. Easwaran et al, unpublished observations). (D) PMEFs from wild-type and Suvar39h1/2 double-null mice expressing TS–GFP and FLAG-tagged HP1β were stained with anti-FLAG antibody and centromeric heterochromatin was visualized with Hoechst 33258. Scale bars, 5 μm.
Figure 4
Recovery of GFP-tagged Dnmt1 after photobleaching indicates de novo loading of Dnmt1 onto chromatin throughout G2. (A) FRAP series of a typical G2 nucleus expressing GMT (green) and RFP–PCNA (red). An area containing a heterochromatic focus was bleached (inset). Scale bar, 5 μm. (B) Recovery curve of the FRAP experiment shown in (A). Half-time of recovery is 11 s. (C) FRAP data of five nuclei were averaged. Mean curve (solid line) and standard deviation (dotted lines) are shown. Half-time of recovery of the mean curve is 11.7 s.
Figure 5
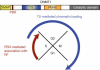
Summary of the role of PBD and TS in controlling the dynamic association of Dnmt1 with chromatin during the cell cycle. PBD directs association of Dnmt1 with RF throughout the S phase and TS mediates association with chromatin, preferentially heterochromatin, during G2 and M.
Similar articles
- Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication.
Estève PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S. Estève PO, et al. Genes Dev. 2006 Nov 15;20(22):3089-103. doi: 10.1101/gad.1463706. Epub 2006 Nov 3. Genes Dev. 2006. PMID: 17085482 Free PMC article. - A role for LSH in facilitating DNA methylation by DNMT1 through enhancing UHRF1 chromatin association.
Han M, Li J, Cao Y, Huang Y, Li W, Zhu H, Zhao Q, Han JJ, Wu Q, Li J, Feng J, Wong J. Han M, et al. Nucleic Acids Res. 2020 Dec 2;48(21):12116-12134. doi: 10.1093/nar/gkaa1003. Nucleic Acids Res. 2020. PMID: 33170271 Free PMC article. - RFTS-dependent negative regulation of Dnmt1 by nucleosome structure and histone tails.
Mishima Y, Brueckner L, Takahashi S, Kawakami T, Arita K, Oka S, Otani J, Hojo H, Shirakawa M, Shinohara A, Watanabe M, Suetake I. Mishima Y, et al. FEBS J. 2017 Oct;284(20):3455-3469. doi: 10.1111/febs.14205. Epub 2017 Sep 11. FEBS J. 2017. PMID: 28834260 - Mammalian cytosine DNA methyltransferase Dnmt1: enzymatic mechanism, novel mechanism-based inhibitors, and RNA-directed DNA methylation.
Svedruzić ZM. Svedruzić ZM. Curr Med Chem. 2008;15(1):92-106. doi: 10.2174/092986708783330700. Curr Med Chem. 2008. PMID: 18220765 Review. - Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns.
Bronner C, Alhosin M, Hamiche A, Mousli M. Bronner C, et al. Genes (Basel). 2019 Jan 18;10(1):65. doi: 10.3390/genes10010065. Genes (Basel). 2019. PMID: 30669400 Free PMC article. Review.
Cited by
- Aberrant regulation of DNA methylation in amyotrophic lateral sclerosis: a new target of disease mechanisms.
Martin LJ, Wong M. Martin LJ, et al. Neurotherapeutics. 2013 Oct;10(4):722-33. doi: 10.1007/s13311-013-0205-6. Neurotherapeutics. 2013. PMID: 23900692 Free PMC article. Review. - DNA methyltransferase 1 mutations and mitochondrial pathology: is mtDNA methylated?
Maresca A, Zaffagnini M, Caporali L, Carelli V, Zanna C. Maresca A, et al. Front Genet. 2015 Mar 12;6:90. doi: 10.3389/fgene.2015.00090. eCollection 2015. Front Genet. 2015. PMID: 25815005 Free PMC article. - Twists and turns of DNA methylation.
Frauer C, Leonhardt H. Frauer C, et al. Proc Natl Acad Sci U S A. 2011 May 31;108(22):8919-20. doi: 10.1073/pnas.1105804108. Epub 2011 May 18. Proc Natl Acad Sci U S A. 2011. PMID: 21593412 Free PMC article. No abstract available. - Distribution of DNA replication proteins in Drosophila cells.
Easwaran HP, Leonhardt H, Cardoso MC. Easwaran HP, et al. BMC Cell Biol. 2007 Oct 15;8:42. doi: 10.1186/1471-2121-8-42. BMC Cell Biol. 2007. PMID: 17937809 Free PMC article. - 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal.
Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. Ghoshal K, et al. Mol Cell Biol. 2005 Jun;25(11):4727-41. doi: 10.1128/MCB.25.11.4727-4741.2005. Mol Cell Biol. 2005. PMID: 15899874 Free PMC article. Retracted.
References
- Bird AP, Wolffe AP (1999) Methylation-induced repression—belts, braces, and chromatin. Cell 99: 451–454 - PubMed
- Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF (1997) Human DNA–(cytosine-5) methyltransferase–PCNA complex as a target for p21WAF1. Science 277: 1996–2000 - PubMed
- Doerfler W (1983) DNA methylation and gene activity. Annu Rev Biochem 52: 93–124 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials