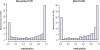DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project - PubMed (original) (raw)
doi: 10.1371/journal.pbio.0020405. Epub 2004 Nov 23.
Thomas Hildmann, Karen L Novik, Jörn Lewin, Jörg Tost, Antony V Cox, T Dan Andrews, Kevin L Howe, Thomas Otto, Alexander Olek, Judith Fischer, Ivo G Gut, Kurt Berlin, Stephan Beck
Affiliations
- PMID: 15550986
- PMCID: PMC529316
- DOI: 10.1371/journal.pbio.0020405
DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project
Vardhman K Rakyan et al. PLoS Biol. 2004 Dec.
Abstract
The Human Epigenome Project aims to identify, catalogue, and interpret genome-wide DNA methylation phenomena. Occurring naturally on cytosine bases at cytosine-guanine dinucleotides, DNA methylation is intimately involved in diverse biological processes and the aetiology of many diseases. Differentially methylated cytosines give rise to distinct profiles, thought to be specific for gene activity, tissue type, and disease state. The identification of such methylation variable positions will significantly improve our understanding of genome biology and our ability to diagnose disease. Here, we report the results of the pilot study for the Human Epigenome Project entailing the methylation analysis of the human major histocompatibility complex. This study involved the development of an integrated pipeline for high-throughput methylation analysis using bisulphite DNA sequencing, discovery of methylation variable positions, epigenotyping by matrix-assisted laser desorption/ionisation mass spectrometry, and development of an integrated public database available at http://www.epigenome.org. Our analysis of DNA methylation levels within the major histocompatibility complex, including regulatory exonic and intronic regions associated with 90 genes in multiple tissues and individuals, reveals a bimodal distribution of methylation profiles (i.e., the vast majority of the analysed regions were either hypo- or hypermethylated), tissue specificity, inter-individual variation, and correlation with independent gene expression data.
Conflict of interest statement
JL, KB, TH, TO, and AO are employees of Epigenomics AG. IGG and SB are members of the scientific advisory board of Epigenomics AG, but do not benefit financially from their involvement in this study. Epigenomics AG has filed for patents based on some of the work described here. Epigenomics AG was a scientific collaborator in the study. The work described here was funded by a grant from the European Union Framework 5 Programme.
Figures
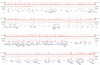
Figure 1. Map of the Human MHC Showing Coverage and Locations of the Bisulphite PCR Amplicons for Which Methylation Data Have Been Generated
Tracks from top to bottom are as follows. (1) CpG content—the proportion of CpGs in 8-kb windows. The expected proportion of CpG dinucletides is 0.04 based on the background base composition of Chromosome 6 (Mungall et al. 2003). (2) Random SNP density in 1,000-bp windows. (3) Location of predicted CpG islands. (4) Bisulphite PCR amplicons. (5) Location of annotated gene structures. Right and left arrows indicate gene structures on the sense and antisense strand, respectively. Official gene symbols are used where available.
Figure 2. Comparison of Methylation Measurements Obtained Using MALDI-MS with Those from ESME Analysis of Directly Sequenced Bisulphite PCR Products
(A) Comparison of methylation measurements obtained by MALDI-MS (x-axis) with ESME-processed data from sequencing (y-axis). Methylation rates at CpGs from forward and reverse sequencing were binned into ten intervals from zero to one using corresponding MALDI-MS measurements at the same CpGs and in the same tissue samples. (B) Comparison of methylation measurements obtained from ESME-processed data (x-axis) with measurements from MALDI-MS (y-axis). Methylation rates from MALDI are binned as in (A), using the corresponding methylation values from sequencing. Red lines show the means of the binned rates; bars show the standard deviations. The overall correlation of the data is 0.887. Data points that are not around a methylation rate of zero or one are covered by few measurements because of the bimodal distribution of methylation measurements.
Figure 3. The HEP Database
(A) We have created a Web-based, ENSEMBL-like genome browser for displaying HEP data that is publicly available at
. The methylation levels calculated by the ESME software are displayed in the form of a matrix. Each matrix contains the data obtained from all the samples of one amplicon. Each colour-coded square (yellow represents 0% methylation, blue represents 100% methylation, and green represents intermediate levels) within the matrix represents one CpG site. Clicking on a square reveals the tissue source of the sample and the level of methylation observed at that particular CpG site. Grey squares indicate CpG sites for which methylation levels could not be determined. Each row of squares represents all the CpG sites for one sample of a particular amplicon, and the samples are grouped by tissue type. The red bar indicates the genomic region analysed. Also shown are chromosome coordinates, CpG islands, SNPs, and ENSEMBL and high-quality, manually curated VEGA transcript information. The HEP database links to the Ensembl genome browser, providing additional information about the region of interest. The example shows amplicons within the SynGAP 1 gene that correspond to regions that were determined to be hypomethylated (second amplicon from the left), hypermethylated (first and fifth amplicons), and heterogeneously methylated (fourth amplicon). Insufficient data were obtained for the third amplicon. (B) By using the zoom function, the user can view the complete DNA sequence for the analysed amplicon.
Figure 4. Bimodal Distribution of DNA Methylation within the Human MHC
(A) Determined by direct sequencing/ESME analysis (based on 86,374 single CpGs in different tissue samples building the median for measurement repetitions). (B) Determined by MALDI-MS (based on 1,019 MALDI measurements).
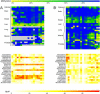
Figure 5. Example of METHANE Output Showing Regions That Display Tissue-Specific Methylation Profiles
The top colour-scale bar refers to the degree of methylation (percent). The bottom colour-scale bar refers to the absolute difference in the methylation level observed between tissues at a given CpG site, and is therefore a measure of the confidence level for a CpG site to be defined as a MVP. (A) The upper matrix represents an amplicon that contains 18 CpG sites within a 386-bp region overlapping exon 3, intron 3, and exon 4 of the complement factor B gene. It is hypomethylated in liver (median methylation is 17%) and hypermethylated in all other tissues examined (median methylation is 100%). The lower matrix shows pairwise comparisons of the methylation values for each CpG site between tissues. (B) The upper matrix represents an amplicon that contains 19 CpG sites within a 550-bp region overlapping exon 3 and intron 3 of the DAXX gene. It is relatively hypomethylated in breast (median methylation is 64%) compared with the other tissues examined (median methylation is 100%). The lower matrix shows pairwise comparisons of the methylation values for each CpG site between tissues.
Figure 6. Example of METHANE Output Showing Regions That Display Inter-Individual Variation of Methylation Profiles
(A) Example of a region that displays significant inter-individual variation, especially in prostate. The matrix represents an amplicon that contains 27 CpG sites within a 527-bp region overlapping the last exon of the CYP21A2 gene. (B) Another example of a region that displays significant inter-individual variation. The matrix represents an amplicon that contains 13 CpG sites within a 453-bp region overlapping the 5′ UTR and exon 1 of the tumour necrosis factor gene.
Figure 7. Comparison of Methylation Values Measured in Five Tissues and Eleven Amplicons Using MALDI-MS and ESME Analysis of Directly Sequenced PCR Products
Each column is a tissue sample, each row a CpG site. Data are ordered in blocks by tissue type and amplicons. Positions of measurements for MALDI-MS (A) correspond to those for ESME analysis (B). The methylation values are colour coded from 0% methylation (yellow) to 100% methylation (blue), with intermediate methylation levels represented by shades of green. White indicates missing measurement values.
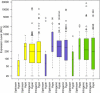
Figure 8. Comparison of DNA Methylation with Gene Expression
Amplicons generated from prostate (yellow), lung (blue), and liver (green) samples were divided into two categories: “upstream” and “intragenic”. The median methylation values for the amplicons were calculated as described in the text, and these were then classified as hypomethylated (median methylation less than 50%) or hypermethylated (median methylation greater than 50%), and plotted against the cDNA microarray expression data available at
(Su et al. 2002). The expression values are expressed as average difference values (ADVs) for each gene. The average difference value is computed using Affymetrix software and is proportional to mRNA content in the sample, with a value of 200 being a conservative cut-off below which a gene can be classified as being not expressed. The average difference values are the mean of 2 or 3 independent experiments. For prostate and liver, the expression levels associated with the hypermethylated upstream amplicons were significantly lower than the expression levels associated with the hypomethylated upstream amplicons (p < 0.0001 for prostate and _p_ < 0.01 for liver). For lung, there was no significant difference between the expression levels associated with the hypermethylated upstream amplicons and those of the hypomethylated upstream amplicons (_p_ > 0.3). There was no correlation between expression and methylation for the intragenic amplicons for any of the three tissues (p > 0.3). The width of the bars is indicative of the number of amplicons in each category: prostate upstream, hypermethylated (n = 9); prostate upstream, hypomethylated (n = 15); prostate intragenic, hypermethylated (n = 109); prostate intragenic, hypomethylated (n = 53); liver upstream, hypermethylated (n = 9); liver upstream, hypomethylated (n = 14); liver intragenic, hypermethylated (n = 115); liver intragenic, hypomethylated (n = 45); lung upstream, hypermethylated (n = 9); lung upstream, hypomethylated (n = 13); lung intragenic, hypermethylated (n = 112); and lung intragenic, hypomethylated (n = 57).
Similar articles
- Future potential of the Human Epigenome Project.
Eckhardt F, Beck S, Gut IG, Berlin K. Eckhardt F, et al. Expert Rev Mol Diagn. 2004 Sep;4(5):609-18. doi: 10.1586/14737159.4.5.609. Expert Rev Mol Diagn. 2004. PMID: 15347255 Review. - DNA methylation: bisulphite modification and analysis.
Patterson K, Molloy L, Qu W, Clark S. Patterson K, et al. J Vis Exp. 2011 Oct 21;(56):3170. doi: 10.3791/3170. J Vis Exp. 2011. PMID: 22042230 Free PMC article. - Epigenomics: genome-wide study of methylation phenomena.
Novik KL, Nimmrich I, Genc B, Maier S, Piepenbrock C, Olek A, Beck S. Novik KL, et al. Curr Issues Mol Biol. 2002 Oct;4(4):111-28. Curr Issues Mol Biol. 2002. PMID: 12432963 Review. - Whole-genome bisulfite sequencing in systemic sclerosis provides novel targets to understand disease pathogenesis.
Lu T, Klein KO, Colmegna I, Lora M, Greenwood CMT, Hudson M. Lu T, et al. BMC Med Genomics. 2019 Oct 24;12(1):144. doi: 10.1186/s12920-019-0602-8. BMC Med Genomics. 2019. PMID: 31651337 Free PMC article. - Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements.
Coolen MW, Statham AL, Gardiner-Garden M, Clark SJ. Coolen MW, et al. Nucleic Acids Res. 2007;35(18):e119. doi: 10.1093/nar/gkm662. Epub 2007 Sep 13. Nucleic Acids Res. 2007. PMID: 17855397 Free PMC article.
Cited by
- Patterns of Genetic And Epigenetic Diversity Across A Range Expansion in The White-Footed Mouse (Peromyscus Leucopus).
Rubi TL, do Prado JR, Knowles LL, Dantzer B. Rubi TL, et al. Integr Org Biol. 2023 Oct 30;5(1):obad038. doi: 10.1093/iob/obad038. eCollection 2023. Integr Org Biol. 2023. PMID: 37942286 Free PMC article. - The Interaction between the Host Genome, Epigenome, and the Gut-Skin Axis Microbiome in Atopic Dermatitis.
Pessôa R, Clissa PB, Sanabani SS. Pessôa R, et al. Int J Mol Sci. 2023 Sep 20;24(18):14322. doi: 10.3390/ijms241814322. Int J Mol Sci. 2023. PMID: 37762624 Free PMC article. Review. - GraphCpG: imputation of single-cell methylomes based on locus-aware neighboring subgraphs.
Deng Y, Tang J, Zhang J, Zou J, Zhu Q, Fan S. Deng Y, et al. Bioinformatics. 2023 Sep 2;39(9):btad533. doi: 10.1093/bioinformatics/btad533. Bioinformatics. 2023. PMID: 37647650 Free PMC article. - The impact of spatial correlation on methylation entropy with application to mouse brain methylome.
Wu X, Choi JM. Wu X, et al. Epigenetics Chromatin. 2023 Feb 4;16(1):5. doi: 10.1186/s13072-023-00479-6. Epigenetics Chromatin. 2023. PMID: 36739438 Free PMC article.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Beck S, Olek A, Walter J. From genomics to epigenomics: A loftier view of life. Nat Biotechnol. 1999;17:1144. - PubMed
- Besterman JM, McLeod R. Targeting gene regulators for cancer therapy: Antisense inhibitors provide new sites for invention. Modern Drug Discov. 2000;April:53–58.
- Bird A. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources