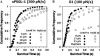Force history dependence of receptor-ligand dissociation - PubMed (original) (raw)
Force history dependence of receptor-ligand dissociation
Bryan T Marshall et al. Biophys J. 2005 Feb.
Abstract
Receptor-ligand bonds that mediate cell adhesion are often subjected to forces that regulate their dissociation via modulating off-rates. Off-rates control how long receptor-ligand bonds last and how much force they withstand. One should therefore be able to determine off-rates from either bond lifetime or unbinding force measurements. However, substantial discrepancies exist between the force dependence of off-rates derived from the two types of measurements even for the same interactions, e.g., selectins dissociating from their ligands, which mediate the tethering and rolling of leukocytes on vascular surfaces during inflammation and immune surveillance. We used atomic force microscopy to measure survival times of P-selectin dissociating from P-selectin glycoprotein ligand 1 or from an antibody in both bond lifetime and unbinding force experiments. By a new method of data analysis, we showed that the discrepancies resulted from the assumption that off-rates were functions of force only. The off-rates derived from forced dissociation data depended not only on force but also on the history of force application. This finding provides a new paradigm for understanding how force regulates receptor-ligand interactions.
Figures
FIGURE 1
AFM experiment. (A) Functionalizing the AFM. The schematic represents a composite of all molecules adsorbed or captured. Adsorbed anti-PSGL-1 mAb PL2 was used to capture sPSGL-1. Anti-P-selectin mAb G1 was adsorbed on a separate tip. P-selectin was reconstituted in a PEI-cushioned lipid bilayer. (B) Force-scan curves. The approach tracing was horizontal (zero mean force) initially but was bent downward when the tip was compressed onto the bilayer. The retraction tracing mirrored the approach tracing until the cantilever returned to the unbent position. After this point, >80% of the retraction tracings leveled off, even though the cantilever continued to retract, indicating the absence of adhesion (top tracing). In <20% of the cases, the retraction tracing was bent upward, indicating a tensile force that was applied to the tip through a molecular bond linked to the bilayer. In the unbinding force experiment, the cantilever was retracted at a constant rate until the tip sprang back to the unbent position (middle tracing). In the bond lifetime experiment, once retracted a predetermined distance, the cantilever was stopped to apply a constant force to the bond (bottom tracing). Unbinding forces were measured along the former loading histories. Bond lifetimes were measured along the constant force portion of the latter loading histories. Survival times were measured along both loading histories (indicated).
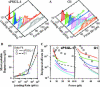
FIGURE 2
Discrepancies between actually measured bond lifetimes and those predicted from unbinding force analysis. (A) Histograms of unbinding forces (>40 measurements for each loading rate) measured at various loading rates for P-selectin interacting with sPSGL-1 (left panel, ∼600 total measurements) and G1 (right panel, ∼450 total measurements). Equations 2 and 3 were fit to each histogram (curves) to determine the most probable unbinding force, _f_m, for that loading rate, _r_f. (B) _f_m was plotted against _r_f in the log scale (points with colors match the histogram colors in (A), which were fitted by Eq. 4 (curves) to evaluate the parameters for each interaction. The best-fit parameters for the P-selectin-sPSGL-1 bonds are _k_1 = 0.0770 s−1, _a_1 = 2.41 nm, _k_2 = 33.6 s−1, and _a_2 = 0.0991 nm. The best-fit parameters for the P-selectin-G1 bonds are: _k_1 = 1.08 s−1, _a_1 = 0.664 nm, _k_2 = 54.7 s−1, and _a_2 = 0.137 nm. (C) Using these parameters and Eq. 3, 1/_k_off was plotted against f (black dashed curves) for P-selectin interacting with sPSGL-1 (left panel) and G1 (right panel). Three closely matched lifetime measures, 〈_t_〉,σ(t), and −1/slope obtained directly from bond lifetime experiments (Marshall et al., 2003; Sarangapani et al., 2004) were averaged and shown for comparison (circles). 1/_k_off versus f data estimated from individual unbinding force histograms according to Eq. 6 (solid curves, colors match those in the histograms) were shown in the force range where bond lifetime data were available for P-selectin dissociating from sPSGL-1 (left panel) and G1 (right panel).
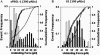
FIGURE 3
Discrepancies between unbinding force histograms and cumulative frequencies actually measured and predicted. Using Eq. 2 and _k_off(f) determined from the bond lifetime data, the cumulative probability _p_c (curves) for a bond (loaded with a constant rate of _r_f = 300 pN/s) to unbind at force not exceeding f and the probability density _p_d (solid bars) for a bond (loaded by the same ramp rate) to unbind at force f were predicted for P-selectin interacting with sPSGL-1 (A) and G1 (B). These were compared to the unbinding forces measured along the same constant-rate loading path and analyzed by histogram (open bars) and cumulative frequency (open circles) for the two interactions.
FIGURE 4
Discrepancies among cumulative frequency versus survival time curves from unbinding force and bond lifetime measurements. Survival times of bonds of P-selectin interacting with sPSGL-1 (A) or G1 (B) were measured from the unbinding force and bond lifetime experiments and analyzed by cumulative frequencies. For the former experiment, bonds were loaded continuously at the indicated constant rates until rupture (solid circles). For the latter experiment, bonds were initially loaded at the same rates to the indicated forces and were then held at those forces. Some of the bonds failed during ramping (solid triangles, squares, and diamonds) whereas others dissociated during holding (open triangles, squares, and diamonds). Only survival times of every third point are shown for clarity. Although two out of three points were skipped to avoid obscuring different symbols, the curve shapes are identical if all data points are included.
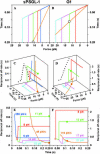
FIGURE 5
3D 1/_k_off curves along various loading histories and their projections on the f-t plane and on the _t_-1/_k_off plane. (A, C, and E) sPSGL-1 data. (B, D, and F) G1 data. (A and B) Loading paths along which the off-rates were measured are depicted for the constant-rate (straight lines) and constant-force (two-segment lines) histories of force application. Note that the value axes are in reverse order so that they can be easily related to the perspective projections from C and D. (C and D) Reciprocal off-rates (1/_k_off, z axis) are plotted as 3D curves in space whose projections on the _f_-t (x-y) plane are the force histories. For the same points on the _f_-t plane, the 1/_k_off values could be very different if evaluated along different force histories, as exemplified by the green and orange solid circles in C and D. The dotted lines show the projections on the _f_-1/_k_off (x-z) plane of the 3D 1/_k_off curves measured along the holding portion of the constant-force force histories, which reveal a catch-slip transitional bond in C and a slip bond in D. (E and F) Projections of the 3D 1/_k_off curves on the _t_-1/_k_off (y-z) plane. Only partial data are shown for clarity. For the 1/_k_off curves determined from the unbinding force data, only those values defined in the same time and force ranges of the bond lifetime data are shown.
Similar articles
- Force modulating dynamic disorder: a physical model of catch-slip bond transitions in receptor-ligand forced dissociation experiments.
Liu F, Ou-Yang ZC. Liu F, et al. Phys Rev E Stat Nonlin Soft Matter Phys. 2006 Nov;74(5 Pt 1):051904. doi: 10.1103/PhysRevE.74.051904. Epub 2006 Nov 6. Phys Rev E Stat Nonlin Soft Matter Phys. 2006. PMID: 17279936 - Force-mediated kinetics of single P-selectin/ligand complexes observed by atomic force microscopy.
Fritz J, Katopodis AG, Kolbinger F, Anselmetti D. Fritz J, et al. Proc Natl Acad Sci U S A. 1998 Oct 13;95(21):12283-8. doi: 10.1073/pnas.95.21.12283. Proc Natl Acad Sci U S A. 1998. PMID: 9770478 Free PMC article. - Molecular dynamics simulation of shear- and stretch-induced dissociation of P-selectin/PSGL-1 complex.
Kang Y, Lü S, Ren P, Huo B, Long M. Kang Y, et al. Biophys J. 2012 Jan 4;102(1):112-20. doi: 10.1016/j.bpj.2011.11.4002. Epub 2012 Jan 3. Biophys J. 2012. PMID: 22225804 Free PMC article. - Theoretical aspects of the biological catch bond.
Prezhdo OV, Pereverzev YV. Prezhdo OV, et al. Acc Chem Res. 2009 Jun 16;42(6):693-703. doi: 10.1021/ar800202z. Acc Chem Res. 2009. PMID: 19331389 Review. - Structure and function of P-selectin glycoprotein ligand-1.
Moore KL. Moore KL. Leuk Lymphoma. 1998 Mar;29(1-2):1-15. doi: 10.3109/10428199809058377. Leuk Lymphoma. 1998. PMID: 9638971 Review.
Cited by
- Probing time-dependent mechanical behaviors of catch bonds based on two-state models.
Chen X, Mao Z, Chen B. Chen X, et al. Sci Rep. 2015 Jan 19;5:7868. doi: 10.1038/srep07868. Sci Rep. 2015. PMID: 25598078 Free PMC article. - Dynamic bonds and their roles in mechanosensing.
Zhu C, Chen Y, Ju LA. Zhu C, et al. Curr Opin Chem Biol. 2019 Dec;53:88-97. doi: 10.1016/j.cbpa.2019.08.005. Epub 2019 Sep 27. Curr Opin Chem Biol. 2019. PMID: 31563813 Free PMC article. Review. - Event-tracking model of adhesion identifies load-bearing bonds in rolling leukocytes.
Pospieszalska MK, Zarbock A, Pickard JE, Ley K. Pospieszalska MK, et al. Microcirculation. 2009 Feb;16(2):115-30. doi: 10.1080/10739680802462792. Epub 2008 Oct 16. Microcirculation. 2009. PMID: 19023690 Free PMC article. - Fast Force Loading Disrupts Molecular Binding Stability in Human and Mouse Cell Adhesions.
Chen Y, Liao J, Yuan Z, Li K, Liu B, Ju LA, Zhu C. Chen Y, et al. Mol Cell Biomech. 2019;16(3):211-223. doi: 10.32604/mcb.2019.07267. Mol Cell Biomech. 2019. PMID: 37063999 Free PMC article. - Effect of viscoelasticity on the analysis of single-molecule force spectroscopy on live cells.
Gupta VK, Neeves KB, Eggleton CD. Gupta VK, et al. Biophys J. 2012 Jul 3;103(1):137-45. doi: 10.1016/j.bpj.2012.05.044. Biophys J. 2012. PMID: 22828340 Free PMC article.
References
- Alon, R., D. A. Hammer, and T. A. Springer. 1995. Lifetime of the P-selectin-carbohydrate bond and its response to tensile force in hydrodynamic flow. Nature. 374:539–542. - PubMed
- Bell, G. I. 1978. Models for the specific adhesion of cells to cells. Science. 200:618–627. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL65631/HL/NHLBI NIH HHS/United States
- R01 AI044902/AI/NIAID NIH HHS/United States
- R01 HL065631/HL/NHLBI NIH HHS/United States
- R21 AI044902/AI/NIAID NIH HHS/United States
- AI44902/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources