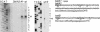Metastability of Helicobacter pylori bab adhesin genes and dynamics in Lewis b antigen binding - PubMed (original) (raw)
Metastability of Helicobacter pylori bab adhesin genes and dynamics in Lewis b antigen binding
Anna Bäckström et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Heterogeneity among Helicobacter pylori strains in gastric epithelial adherence is postulated to contribute to pathogen fitness in the physiologically diverse human population. H. pylori adherence to ABO and Lewis b (Leb) blood group antigens in the human stomach is mediated by the blood group antigen-binding adhesin BabA. Approximately 70% of Swedish and U.S. H. pylori clinical isolates exhibit Leb binding, but here we show that the babA gene is present in each of 10 Leb-nonbinding strains. Fluorescence microscopy identified occasional bacterial cells with a Leb-binding phenotype in populations of Leb-nonbinding strains. Thus, nonbinding seemed to be a metastable phenotype. To model metastable transition into the virulence-associated Leb-binding mode, Leb-binding clones were isolated from nonadherent strains by panning with Leb-magnetic beads and characterized. Strain 17875 has two babA genes, babA1 (silent) and babA2 (expressed). We found that a babA2-cam derivative of strain 17875 regained Leb binding by recombination of the formerly silent babA1 gene into the expressed and partially homologous babB locus. The chimeric BabB/A adhesin binds Leb with an affinity similar to that of wild-type BabA adhesin, but its expression level was lower and was subject to phase variation through slipped-strand mispairing. Equivalent results were obtained with strain NCTC11638. We propose that adhesin metastability and heterogeneity contributes to bacterial fitness and results in some clones having potential for periodic activation and deactivation of virulence appropriate for intensity of the host response to infection.
Figures
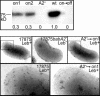
Fig. 1.
Leb binding of OFF strain and ON variants. (Left) Fluorescence microscopy showing occasional Leb-binding bacterial cells detected with Cy-3-streptavidin and nonbinding cells with FITC. Hence, a bacterial cell with an OFF→ ON Leb-binding phenotype is detected by yellowish staining among green nonbinding cells of 17875Δ_babA2_::cam. (Center) For affinity analyses according to Scatchard, the 125I-labeled Leb conjugate was diluted with unlabeled Leb conjugate and binding affinity was measured. Under these conditions, wild-type strain 17875 (•), A1on1 (▪), and A1on2 (▴) exhibited affinity constants of 1.5 × 10-11, 1.0 × 10-11, and 0.74 × 10-11 M-1, respectively. (Right) The full Leb-binding capacity was calculated for strains 17875, A1on1, and A1on2. The binding for A1on1 and A1on2 is shown relative to that of wild-type strain 17875.
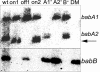
Fig. 2.
Immunoblot and ImmunoGold electron microscopy analysis of OFF strain and ON variants. (Upper) Immunoblot analysis of whole-cell extracts by using Ak253 BabA antibodies. Lanes: 1, A1on1 (on1); 2, A1on2 (on2); 3, 17875Δ_babA2_::cam (A2-); 4, wild-type 17875 (wt); 5, off phase shift of A1on1 (ON→ OFF). The relative amount of adhesin expressed in A1on1 and A1on2 was 0.3 as compared with the wild-type strain 17875 (1.0). (Lower) Electron micrographs of H. pylori. BabA and the BabB/A chimeric adhesin expressed on the bacterial surface is visualized by immunogold labeling with anti-BabA and 10-nm gold particles. Gold particles on five electron micrographs of each strain were counted, and the ratio of the wild-type 17875 to A1on1 was 0.25.
Fig. 3.
Southern blot analysis of the isolated A1on clones. Genomic DNA isolated from strains 17875 wild type (wt); A1on1 (on1); A1off (off1); A1on2 (on2), 17875Δ_babA1_::cam (A1-); 17875Δ_babA2_::cam (A2-); 17875Δ_babB_::cam (B-); 17875Δ_babA2_::camΔ_babA1_::kan (DM), digested with Hin_dIII. (Upper) Hybridization to DNA of the 17875Δ_babA1::cam strain identified a babA2 specific fragment of 1,000 bp, whereas a 3,000-bp babA1_-specific fragment was present in 17875Δ_babA2::cam. Both fragments are present in parent strain 17875 (wt). In the A1on clones, the _babA1_-specific fragment and an additional fragment of ≈900 bp were detected. (Lower) A 2,300-bp, babB_-specific fragment was present in all strains except for the A1on clones and the babB knockout mutant, 17875Δ_babB::cam (B-).
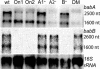
Fig. 4.
Northern blot analysis of total RNA isolated from strains 17875 wild type (wt), A1on1 (On1), A1on2 (On2), 17875Δ_babA1_::cam (A1-), 17875Δ_babA2_::cam (A2-), 17875Δ_babB_::cam (B-), and 17875Δ_babA2_::camΔ_babA1_::kan (DM) and hybridized with a babA-specific probe. (Top) A specific 2,500-nt transcript that corresponds to the size of the babA gene in wild-type 17875 and 17875Δ_babA1::cam mutant cells but not in the 17875Δ_babA2::cam mutant or the double mutant 17875Δ_babA2_::camΔ_babA1_::kan. (Middle) The same filter hybridized with a babB probe identified a 2,600-nt babB transcript. A 1,600-nt transcript was present together with both the babA and the babB transcripts. (Bottom) The amount of RNA in each lane is visualized by hybridization with a 16S rRNA probe. The expression of the babA mRNA (Top) in A1on1 and A1on2 was quantified in relation to expression in strain 17875, for which the values from the 16S hybridization were used as references: 17875, 1.0; A1on1, 0.32; A1on2, 0.29.
Fig. 5.
Primer extension analysis. (Left) Analysis of the 5′ end of the babA2 transcripts. Total RNA was used for the primer extension with 32P-end-labeled oligonucleotide primers. babA2 extension products were analyzed from strain 17875 wild type (wt), 17875Δ_babA1_::cam (A1-), 17875Δ_babA2_::cam (A2-), and 17875Δ_babA2_::camΔ_babA1_::kan (DM). (Center) Primer extension analysis of the babB operon. Total RNA was used for the primer extension with 32P-end-labeled oligonucleotide primer. babB extension products were analyzed from 17875 wild type and 17875Δ_babB_::cam (B-). Sequence analysis of DNA corresponds to the respective upstream region and serves as a reference. The sequence analysis was performed with the same primer as used for the primer extension. (Right) The -10 and -35 promoter sequences are marked in the respective DNA sequences.
Similar articles
- Helicobacter pylori Strains from Duodenal Ulcer Patients Exhibit Mixed babA/B Genotypes with Low Levels of BabA Adhesin and Lewis b Binding.
Saberi S, Schmidt A, Eybpoosh S, Esmaili M, Talebkhan Y, Mohajerani N, Oghalaie A, Eshagh Hosseini M, Mohagheghi MA, Bugaytova J, Borén T, Mohammadi M. Saberi S, et al. Dig Dis Sci. 2016 Oct;61(10):2868-2877. doi: 10.1007/s10620-016-4217-z. Epub 2016 Jun 18. Dig Dis Sci. 2016. PMID: 27318698 - Dynamics of Lewis b binding and sequence variation of the babA adhesin gene during chronic Helicobacter pylori infection in humans.
Nell S, Kennemann L, Schwarz S, Josenhans C, Suerbaum S. Nell S, et al. mBio. 2014 Dec 16;5(6):e02281-14. doi: 10.1128/mBio.02281-14. mBio. 2014. PMID: 25516619 Free PMC article. - Key importance of the Helicobacter pylori adherence factor blood group antigen binding adhesin during chronic gastric inflammation.
Prinz C, Schöniger M, Rad R, Becker I, Keiditsch E, Wagenpfeil S, Classen M, Rösch T, Schepp W, Gerhard M. Prinz C, et al. Cancer Res. 2001 Mar 1;61(5):1903-9. Cancer Res. 2001. PMID: 11280745 - Roles of Helicobacter pylori BabA in gastroduodenal pathogenesis.
Yamaoka Y. Yamaoka Y. World J Gastroenterol. 2008 Jul 21;14(27):4265-72. doi: 10.3748/wjg.14.4265. World J Gastroenterol. 2008. PMID: 18666312 Free PMC article. Review. - Adherence properties of Helicobacter pylori: impact on pathogenesis and adaptation to the host.
Odenbreit S. Odenbreit S. Int J Med Microbiol. 2005 Sep;295(5):317-24. doi: 10.1016/j.ijmm.2005.06.003. Int J Med Microbiol. 2005. PMID: 16173498 Review.
Cited by
- Helicobacter pylori Strains from Duodenal Ulcer Patients Exhibit Mixed babA/B Genotypes with Low Levels of BabA Adhesin and Lewis b Binding.
Saberi S, Schmidt A, Eybpoosh S, Esmaili M, Talebkhan Y, Mohajerani N, Oghalaie A, Eshagh Hosseini M, Mohagheghi MA, Bugaytova J, Borén T, Mohammadi M. Saberi S, et al. Dig Dis Sci. 2016 Oct;61(10):2868-2877. doi: 10.1007/s10620-016-4217-z. Epub 2016 Jun 18. Dig Dis Sci. 2016. PMID: 27318698 - Biochemical and functional characterization of Helicobacter pylori vesicles.
Olofsson A, Vallström A, Petzold K, Tegtmeyer N, Schleucher J, Carlsson S, Haas R, Backert S, Wai SN, Gröbner G, Arnqvist A. Olofsson A, et al. Mol Microbiol. 2010 Sep;77(6):1539-55. doi: 10.1111/j.1365-2958.2010.07307.x. Epub 2010 Aug 5. Mol Microbiol. 2010. PMID: 20659286 Free PMC article. - From array-based hybridization of Helicobacter pylori isolates to the complete genome sequence of an isolate associated with MALT lymphoma.
Thiberge JM, Boursaux-Eude C, Lehours P, Dillies MA, Creno S, Coppée JY, Rouy Z, Lajus A, Ma L, Burucoa C, Ruskoné-Foumestraux A, Courillon-Mallet A, De Reuse H, Boneca IG, Lamarque D, Mégraud F, Delchier JC, Médigue C, Bouchier C, Labigne A, Raymond J. Thiberge JM, et al. BMC Genomics. 2010 Jun 10;11:368. doi: 10.1186/1471-2164-11-368. BMC Genomics. 2010. PMID: 20537153 Free PMC article. - Helicobacter pylori's unconventional role in health and disease.
Dorer MS, Talarico S, Salama NR. Dorer MS, et al. PLoS Pathog. 2009 Oct;5(10):e1000544. doi: 10.1371/journal.ppat.1000544. Epub 2009 Oct 26. PLoS Pathog. 2009. PMID: 19855816 Free PMC article. Review. - Gain and loss of multiple genes during the evolution of Helicobacter pylori.
Gressmann H, Linz B, Ghai R, Pleissner KP, Schlapbach R, Yamaoka Y, Kraft C, Suerbaum S, Meyer TF, Achtman M. Gressmann H, et al. PLoS Genet. 2005 Oct;1(4):e43. doi: 10.1371/journal.pgen.0010043. PLoS Genet. 2005. PMID: 16217547 Free PMC article.
References
- Cover, T., Berg, D. E., Blaser, M. J. & Mobley, H. (2001) in Principles of Bacterial Pathogenesis, ed. Groisman, E. A. (Academic, New York), pp. 509-558.
- Borén, T., Falk, P., Roth, K. A., Larson, G. & Normark, S. (1993) Science 262, 1892-1895. - PubMed
- Aspholm-Hurtig, M., Dailide, G., Lahmann, M., Kalia, A., Ilver, D., Roche, N., Vikström, S., Sjöstrom, R., Lindén, S., Bäckström, A., et al. (2004) Science 305, 519-522. - PubMed
- Ilver, D., Arnqvist, A., Ögren, J., Frick, I.-M., Kersulyte, D., Incecik, E. T., Berg, D. E., Covacci, A., Engstrand, L. & Borén, T. (1998) Science 279, 373-377. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 A1316608/PHS HHS/United States
- R0 DK63041/DK/NIDDK NIH HHS/United States
- R01 DK063041/DK/NIDDK NIH HHS/United States
- P30 DK52474/DK/NIDDK NIH HHS/United States
- R01 HD053727/HD/NICHD NIH HHS/United States
- R01 DK53727/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources