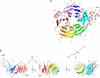Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex - PubMed (original) (raw)
Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex
Ian C Berke et al. J Cell Biol. 2004.
Abstract
Nucleocytoplasmic transport occurs through nuclear pore complexes (NPCs) whose complex architecture is generated from a set of only approximately 30 proteins, termed nucleoporins. Here, we explore the domain structure of Nup133, a nucleoporin in a conserved NPC subcomplex that is crucial for NPC biogenesis and is believed to form part of the NPC scaffold. We show that human Nup133 contains two domains: a COOH-terminal domain responsible for its interaction with its subcomplex through Nup107; and an NH2-terminal domain whose crystal structure reveals a seven-bladed beta-propeller. The surface properties and conservation of the Nup133 beta-propeller suggest it may mediate multiple interactions with other proteins. Other beta-propellers are predicted in a third of all nucleoporins. These and several other repeat-based motifs appear to be major elements of nucleoporins, indicating a level of structural repetition that may conceptually simplify the assembly and disassembly of this huge protein complex.
Figures
Figure 1.
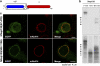
Domain structure of Nup133. (a) Two domains were predicted, with the NTD having helical and sheet content, and the CTD being all helical. The construct used for crystallization is boxed. (b) In vitro binding of hNup133 domains to recombinant GST-hNup107. Top and bottom panels show the bound and unbound fractions of various [35S]methionine-labeled Nup133 translation products incubated with recombinant GST-Nup107 immobilized on affinity resin. (c) Localization of EGFP-tagged hNup133 domains in HeLa cells. The CTD shows punctate rim staining that overlaps with the mAb414 signal. The NTD shows diffuse EGFP-signal, with a weak concentration at the nuclear rim.
Figure 2.
Architecture of hNup133 NTD. (a) Ribbon representation of hNup133 NTD β-propeller, top view. The four strands of each blade are labeled A–D. Blades are labeled 1–7 and colored red through purple with helical insertions shown in pink. The disordered loops are represented as transparent tubes. (b) Side views of the propeller related ∼120° to each other. Figures were prepared with Dino (
) and Povray (
).
Figure 3.
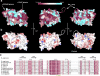
Structural alignment of the seven blades in the Nup133 β-propeller. (a and b) Superposition of 21 Cα positions from each blade calculated by the program MultiProt (
http://bioinfo3d.cs.tau.ac.il/MultiProt/
). Gray spheres mark the aligned Cα atoms from blade 3. Strand A lines the inner channel of the propeller and is roughly parallel to the pseudo-sevenfold axis. Blade 2 (orange), blade 5 (cyan), and blade 7 (purple) contain insertions. (c) Structure-based sequence alignment of Nup133 NTD blades. Conserved hydrophobic positions are shaded yellow. Clathrin NTD propeller blades are shown for comparison. The sequence is shown in Joy formatting (
http://www-cryst.bioc.cam.ac.uk/\~joy/
). β-Strands are shown in blue, 310 helices are shown in maroon, capitalized residues are solvent inaccessible, bold indicates a sidechain-backbone amide H-bond, underline indicates sidechain-backbone carbonyl H-bond, and italics indicates a residue with positive phi values. Gray residues were not built in the model. Bracketed numbers refer to residues not depicted.
Figure 4.
Conserved features of the Nup133 NTD propeller. (a) Surface representation showing conservation of residues on the propeller sides. Each view is an ∼120° rotation about the pseudo-sevenfold axis beginning from the NH2 and COOH termini in blade 7 and is the same as in Fig. 2 b. The tube marks the region of the disordered DA34 loop. Conservation scores were calculated using the ConSurf Server (
) and colored maroon (most conserved) to cyan (most variable). (b) Electrostatic potential of the Nup133 β-propeller. Orientation is as in part a. Red indicates negative regions of the potential and blue indicates positive regions. (c) Sequence alignment of the DA34 loop across representative metazoans. Position of the conserved protein kinase A consensus site is boxed and potential phosphorylated residues are marked by yellow stars. Green triangles mark conserved hydrophobic positions.
Similar articles
- Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex.
Boehmer T, Jeudy S, Berke IC, Schwartz TU. Boehmer T, et al. Mol Cell. 2008 Jun 20;30(6):721-31. doi: 10.1016/j.molcel.2008.04.022. Mol Cell. 2008. PMID: 18570875 Free PMC article. - Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex.
Kim SJ, Fernandez-Martinez J, Sampathkumar P, Martel A, Matsui T, Tsuruta H, Weiss TM, Shi Y, Markina-Inarrairaegui A, Bonanno JB, Sauder JM, Burley SK, Chait BT, Almo SC, Rout MP, Sali A. Kim SJ, et al. Mol Cell Proteomics. 2014 Nov;13(11):2911-26. doi: 10.1074/mcp.M114.040915. Epub 2014 Aug 19. Mol Cell Proteomics. 2014. PMID: 25139911 Free PMC article. - Analysis of the initiation of nuclear pore assembly by ectopically targeting nucleoporins to chromatin.
Schwartz M, Travesa A, Martell SW, Forbes DJ. Schwartz M, et al. Nucleus. 2015;6(1):40-54. doi: 10.1080/19491034.2015.1004260. Nucleus. 2015. PMID: 25602437 Free PMC article. - Structure and Assembly of the Nuclear Pore Complex.
Hampoelz B, Andres-Pons A, Kastritis P, Beck M. Hampoelz B, et al. Annu Rev Biophys. 2019 May 6;48:515-536. doi: 10.1146/annurev-biophys-052118-115308. Epub 2019 Apr 3. Annu Rev Biophys. 2019. PMID: 30943044 Review. - Functional architecture of the nuclear pore complex.
Grossman E, Medalia O, Zwerger M. Grossman E, et al. Annu Rev Biophys. 2012;41:557-84. doi: 10.1146/annurev-biophys-050511-102328. Annu Rev Biophys. 2012. PMID: 22577827 Review.
Cited by
- Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element.
Whittle JRR, Schwartz TU. Whittle JRR, et al. J Biol Chem. 2009 Oct 9;284(41):28442-28452. doi: 10.1074/jbc.M109.023580. Epub 2009 Aug 11. J Biol Chem. 2009. PMID: 19674973 Free PMC article. - Dunking into the Lipid Bilayer: How Direct Membrane Binding of Nucleoporins Can Contribute to Nuclear Pore Complex Structure and Assembly.
Hamed M, Antonin W. Hamed M, et al. Cells. 2021 Dec 20;10(12):3601. doi: 10.3390/cells10123601. Cells. 2021. PMID: 34944108 Free PMC article. Review. - A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase.
Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V. Bolhy S, et al. J Cell Biol. 2011 Mar 7;192(5):855-71. doi: 10.1083/jcb.201007118. J Cell Biol. 2011. PMID: 21383080 Free PMC article. - Compositionally distinct nuclear pore complexes of functionally distinct dimorphic nuclei in the ciliate Tetrahymena.
Iwamoto M, Osakada H, Mori C, Fukuda Y, Nagao K, Obuse C, Hiraoka Y, Haraguchi T. Iwamoto M, et al. J Cell Sci. 2017 May 15;130(10):1822-1834. doi: 10.1242/jcs.199398. Epub 2017 Apr 6. J Cell Sci. 2017. PMID: 28386019 Free PMC article. - Structural and functional studies of the 252 kDa nucleoporin ELYS reveal distinct roles for its three tethered domains.
Bilokapic S, Schwartz TU. Bilokapic S, et al. Structure. 2013 Apr 2;21(4):572-80. doi: 10.1016/j.str.2013.02.006. Epub 2013 Mar 14. Structure. 2013. PMID: 23499022 Free PMC article.
References
- Andrade, M.A., C. Perez-Iratxeta, and C.P. Ponting. 2001. Protein repeats: structures, functions, and evolution. J. Struct. Biol. 134:117–131. - PubMed
- Brunger, A.T., P.D. Adams, G.M. Clore, W.L. DeLano, P. Gros, R.W. Grosse-Kunstleve, J.S. Jiang, J. Kuszewski, M. Nilges, N.S. Pannu, et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous