A mutation in a novel ATP-dependent Lon protease gene in a kindred with mild mental retardation - PubMed (original) (raw)
Comparative Study
A mutation in a novel ATP-dependent Lon protease gene in a kindred with mild mental retardation
Joseph J Higgins et al. Neurology. 2004.
Abstract
Background: Identifying the genetic factors that contribute to memory and learning is limited by the complexity of brain development and the lack of suitable human models for mild disorders of cognition.
Methods: Previously, a disease locus was mapped for a mild type of nonsyndromic mental retardation (IQ between 50 and 70) to a 4.2-MB interval on chromosome 3p25-pter in a large kindred. The genes and transcripts within the candidate region were systematically analyzed for mutations by single-strand polymorphism analysis and DNA sequencing.
Results: A nonsense mutation causing a premature stop codon in a novel gene (cereblon; CRBN) was identified that encodes for an ATP-dependent Lon protease. The predicted protein sequence is highly conserved across species, and it belongs to a family of proteins that selectively degrade short-lived polypeptides and regulate mitochondrial replication and transcription. One member of the Lon-containing protein family is regionally expressed in the human hippocampus, an important neuroanatomic region that is involved in long-term potentiation and learning. The mutation in the CRBN gene described interrupts an N-myristoylation site and eliminates a casein kinase II phosphorylation site at the C terminus.
Conclusions: A gene on chromosome 3p that is associated with mild mental retardation in a large kindred is reported. This finding implicates a role for the ATP-dependent degradation of proteins in memory and learning.
Figures
Figure 1
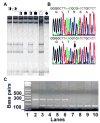
Identification of the 1,274C_→_T mutation in CRBN _exon 11. (A) Representative sample of the single-strand conformational polymorphism (SSCP) mobility shifts in one family branch with mild autosomal recessive nonsyndromic mental retardation. SSCP was performed on the 331-bp PCR products of exon 11, including its 5_′ _and 3_′ splice sites. The mobility shift patterns co-segregated with the affection status and correlated with the results of linkage analysis reported previously. Affected individuals (filled symbols) have two upper bands and one lower band. Carriers (partially filled symbols) have six bands including two upper bands, two middle bands, and two lower bands. Noncarriers (open symbols) have one upper band, two middle bands, and two lower bands. The SSCP pattern of the noncarrier was the same as the patterns in 200 normal individuals (400 chromosomes). (B) Direct sequencing of PCR products. (Top) The CRBN sequence between nucleotide (nt) 1,263 and 1,284 from a normal individual (noncarrier). (Bottom) A 1,274C_→_T substitution in an affected individual in the family. Below the nucleotide sequence is the amino acid sequence. The mutation (arrow) results in a stop codon at the arginine (R) residue at codon 419 and truncates the CRBN protein (R419X). (C) A 2% agarose/0.1% ethidium bromide gel showing the HpyCH4IV restriction enzyme analysis of CRBN exon 11. Lane 1 is a 100-bp ladder. The PCR product of a normal individual (lane 2) is not cleaved by HpyCH4IV and shows a single band representing the 331-bp wild-type alleles. Carriers show two bands representing the 331-bp wild-type allele and the cleaved mutant allele (164-bp, 167-bp) (lanes 3 to 6). The mutation creates a restriction site so that the amplified 331-bp fragment is cleaved in half (164-bp, 167-bp) (lanes 7 to 10).
Figure 2
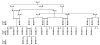
Segregation of the CRBN mutation using HpyCH4IV restriction enzyme digestion of exon 11 in an abridged family pedigree. As in figure 1C, a single 331-bp band represents the wild-type allele and is homozygous in Individuals 2, 8, 10, 19, 24, 26, 30, and 31. Carriers (Individuals 1, 3, 4, 5, 6, 7, 9, 12, 15, 16, 20, 22, 27, 28, and 32) show two bands representing the 331-bp wild-type allele and the cleaved mutant allele (164-bp, 167-bp). The mutation creates a restriction site so that the amplified 331-bp fragment is cleaved in half (164-bp, 167-bp) and is shown as a single lower band in affected Individuals 11, 13, 14, 17, 18, 21, 23, 25, and 29. Filled circles (females) and squares (males) represent affected individuals with mental retardation. Unaffected individuals are indicated by open symbols. Consanguineous marriages are indicated by double bars joining the individuals. For confidentiality reasons, the order and the sex of at-risk individuals are changed.
Figure 3
Alignment of the amino acid sequences from different species with similarities to the CRBN protein. Low consensus alignments are in gray and neutral consensus alignments are in black. The underlined sequences show the Lon domain from amino acid residues 81 to 317 of the predicted protein for human CRBN and the homologous proteins for mice and rats. Prosite predictions for the casein kinase II phosphorylation site (filled circles), myristoylation sites (open circles), protein kinase C phosphorylation site (filled squares), and a glycosylation site (open squares) are shown. The arrow shows the position of the stop codon (R419X) that interrupts an N_-myristoylation site and eliminates a casein kinase II phosphorylation site._
Similar articles
- Primary function analysis of human mental retardation related gene CRBN.
Xin W, Xiaohua N, Peilin C, Xin C, Yaqiong S, Qihan W. Xin W, et al. Mol Biol Rep. 2008 Jun;35(2):251-6. doi: 10.1007/s11033-007-9077-3. Epub 2007 Mar 23. Mol Biol Rep. 2008. PMID: 17380424 - Rescue of Learning and Memory Deficits in the Human Nonsyndromic Intellectual Disability Cereblon Knock-Out Mouse Model by Targeting the AMP-Activated Protein Kinase-mTORC1 Translational Pathway.
Bavley CC, Rice RC, Fischer DK, Fakira AK, Byrne M, Kosovsky M, Rizzo BK, Del Prete D, Alaedini A, Morón JA, Higgins JJ, D'Adamio L, Rajadhyaksha AM. Bavley CC, et al. J Neurosci. 2018 Mar 14;38(11):2780-2795. doi: 10.1523/JNEUROSCI.0599-17.2018. Epub 2018 Feb 19. J Neurosci. 2018. PMID: 29459374 Free PMC article. - A missense mutation in the CRBN gene that segregates with intellectual disability and self-mutilating behaviour in a consanguineous Saudi family.
Sheereen A, Alaamery M, Bawazeer S, Al Yafee Y, Massadeh S, Eyaid W. Sheereen A, et al. J Med Genet. 2017 Apr;54(4):236-240. doi: 10.1136/jmedgenet-2016-104117. Epub 2017 Jan 31. J Med Genet. 2017. PMID: 28143899 Free PMC article. - Genetics of autosomal recessive non-syndromic mental retardation: recent advances.
Basel-Vanagaite L. Basel-Vanagaite L. Clin Genet. 2007 Sep;72(3):167-74. doi: 10.1111/j.1399-0004.2007.00881.x. Clin Genet. 2007. PMID: 17718851 Review. - Fruit flies and intellectual disability.
Bolduc FV, Tully T. Bolduc FV, et al. Fly (Austin). 2009 Jan-Mar;3(1):91-104. doi: 10.4161/fly.3.1.7812. Epub 2009 Jan 12. Fly (Austin). 2009. PMID: 19182539 Free PMC article. Review.
Cited by
- Ablation of CRBN induces loss of type I collagen and SCH in mouse skin by fibroblast senescence via the p38 MAPK pathway.
Jeon S, Yoon YS, Kim HK, Han J, Lee KM, Seol JE, Cho SK, Park CS. Jeon S, et al. Aging (Albany NY). 2021 Mar 3;13(5):6406-6419. doi: 10.18632/aging.202744. Epub 2021 Mar 3. Aging (Albany NY). 2021. PMID: 33658395 Free PMC article. - Hypnotic effect of thalidomide is independent of teratogenic ubiquitin/proteasome pathway.
Hirose Y, Kitazono T, Sezaki M, Abe M, Sakimura K, Funato H, Handa H, Vogt KE, Yanagisawa M. Hirose Y, et al. Proc Natl Acad Sci U S A. 2020 Sep 15;117(37):23106-23112. doi: 10.1073/pnas.1917701117. Epub 2020 Aug 26. Proc Natl Acad Sci U S A. 2020. PMID: 32848052 Free PMC article. - Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability.
Riazuddin S, Hussain M, Razzaq A, Iqbal Z, Shahzad M, Polla DL, Song Y, van Beusekom E, Khan AA, Tomas-Roca L, Rashid M, Zahoor MY, Wissink-Lindhout WM, Basra MAR, Ansar M, Agha Z, van Heeswijk K, Rasheed F, Van de Vorst M, Veltman JA, Gilissen C, Akram J, Kleefstra T, Assir MZ; UK10K; Grozeva D, Carss K, Raymond FL, O'Connor TD, Riazuddin SA, Khan SN, Ahmed ZM, de Brouwer APM, van Bokhoven H, Riazuddin S. Riazuddin S, et al. Mol Psychiatry. 2017 Nov;22(11):1604-1614. doi: 10.1038/mp.2016.109. Epub 2016 Jul 26. Mol Psychiatry. 2017. PMID: 27457812 Free PMC article. - Cereblon negatively regulates TLR4 signaling through the attenuation of ubiquitination of TRAF6.
Min Y, Wi SM, Kang JA, Yang T, Park CS, Park SG, Chung S, Shim JH, Chun E, Lee KY. Min Y, et al. Cell Death Dis. 2016 Jul 28;7(7):e2313. doi: 10.1038/cddis.2016.226. Cell Death Dis. 2016. PMID: 27468689 Free PMC article. - The CC2D1A, a member of a new gene family with C2 domains, is involved in autosomal recessive non-syndromic mental retardation.
Basel-Vanagaite L, Attia R, Yahav M, Ferland RJ, Anteki L, Walsh CA, Olender T, Straussberg R, Magal N, Taub E, Drasinover V, Alkelai A, Bercovich D, Rechavi G, Simon AJ, Shohat M. Basel-Vanagaite L, et al. J Med Genet. 2006 Mar;43(3):203-10. doi: 10.1136/jmg.2005.035709. Epub 2005 Jul 20. J Med Genet. 2006. PMID: 16033914 Free PMC article.
References
- Massey PS, McDermott S. State-specific rates of mental retardation—United States, 1993. MMWR. 1996;45:61–65. - PubMed
- Curry CJ, Stevenson RE, Aughton D, et al. Evaluation of mental retardation: recommendations of a Consensus Conference: American College of Medical Genetics. Am J Med Genet. 1997;72:468–477. - PubMed
- Wright SW, Tarjan G, Eyer L. Investigation of families with two or more mentally defective siblings; clinical observations. Am J Dis Child. 1959;97:445–463. - PubMed
- Priest JH, Thuline HC, Laveck GD, Jarvis DB. An approach to genetic factors in mental retardation. Studies of families containing at least two siblings admitted to a state institution for the retarded. Am J Ment Defic. 1961;66:42–50. - PubMed
- Bartley JA, Hall BD. Mental retardation and multiple congenital anomalies of unknown etiology: frequency of occurrence in similarly affected sibs of the proband. Birth Defects. 1978;14:127–137. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous