Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity - PubMed (original) (raw)
Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity
Brian H Harcourt et al. J Virol. 2004 Dec.
Abstract
Gene 1 of the coronavirus associated with severe acute respiratory syndrome (SARS) encodes replicase polyproteins that are predicted to be processed into 16 nonstructural proteins (nsps 1 to 16) by two viral proteases, a papain-like protease (PLpro) and a 3C-like protease (3CLpro). Here, we identify SARS coronavirus amino-terminal replicase products nsp1, nsp2, and nsp3 and describe trans-cleavage assays that characterize the protease activity required to generate these products. We generated polyclonal antisera to glutathione S-transferase-replicase fusion proteins and used the antisera to detect replicase intermediates and products in pulse-chase experiments. We found that nsp1 (p20) is rapidly processed from the replicase polyprotein. In contrast, processing at the nsp2/3 site is less efficient, since a approximately 300-kDa intermediate (NSP2-3) is detected, but ultimately nsp2 (p71) and nsp3 (p213) are generated. We found that SARS coronavirus replicase products can be detected by 4 h postinfection in the cytoplasm of infected cells and that nsps 1 to 3 colocalize with newly synthesized viral RNA in punctate, perinuclear sites consistent with their predicted role in viral RNA synthesis. To determine if PLpro is responsible for processing these products, we cloned and expressed the PLpro domain and the predicted substrates and established PLpro trans-cleavage assays. We found that the PLpro domain is sufficient for processing the predicted nsp1/2 and nsp2/3 sites. Interestingly, expression of an extended region of PLpro that includes the downstream hydrophobic domain was required for processing at the predicted nsp3/4 site. We found that the hydrophobic domain is inserted into membranes and that the lumenal domain is glycosylated at asparagine residues 2249 and 2252. Thus, the hydrophobic domain may anchor the replication complex to intracellular membranes. These studies revealed that PLpro can cleave in trans at the three predicted cleavage sites and that it requires membrane association to process the nsp3/4 cleavage site.
Figures
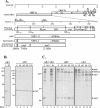
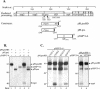
FIG. 1.
Identification of SARS coronavirus ORF1a intermediate NSP2-3 and products nsp1, nsp2, and nsp3 by pulse-chase analysis. (A) Schematic diagram illustrating the SARS coronavirus open reading frames (ORFs), the predicted processing of polyprotein 1a (pp1a) to nonstructural protein (nsp) products, and regions used to generate anti-R1, anti-R2, and anti-R3 sera. The papain-like proteinase domain (PLpro), hydrophobic domain (HD), and picornavirus 3C-like proteinase domain (3CLpro) are designated. (B) Detection of nsp1 by immunoprecipitation with anti-R1 serum from SARS coronavirus-infected (I) and mock-infected (M) cells radiolabeled with [35S]methionine for 60 min from 21 to 22 h postinfection. For the pulse-chase analysis, SARS coronavirus-infected Vero E6 cells were pulse-labeled with [35S]methionine from 21.5 to 22.0 h postinfection. The labeling medium was then replaced with complete DMEM. Cells were lysed at the chase times indicated in lanes 3 to 7, and total lysates were subjected to immunoprecipitation with anti-R1 serum. Products were analyzed on an SDS-12.0% polyacrylamide gel and subjected to autoradiography. (C) Detection of NSP2-3 intermediate and processed products nsp2 and nsp3 by immunoprecipitation with anti-R2 and anti-R3 sera. SARS coronavirus-infected, radiolabeled cell lysates were prepared as above, and products were analyzed on an SDS-8.0% polyacrylamide gel and subjected to autoradiography. The positions of molecular size markers are shown on the left of each gel (in kilodaltons).
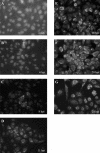
FIG. 2.
Detection of SARS coronavirus replicase products by indirect immunofluorescent staining. Vero E6 cells were infected with 1,000 TCID50 of SARS coronavirus Urbani, fixed and permeabilized at 4, 8, 11, 16, 20, or 24 h postinfection, and stained with anti-R3 and fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin as described in Materials and Methods.
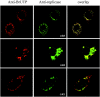
FIG. 3.
Colocalization of SARS coronavirus replicase products with de novo-synthesized viral RNA. SARS coronavirus-infected cells were transfected with Lipofectamine containing BrUTP at 21 h postinfection. Cells were treated with actinomycin D (5 μg/ml) to block cellular mRNA synthesis. The cells were double stained with rabbit polyclonal antisera against nsp1 (anti-R1), nsp2 (anti-R2), or nsp3 (anti-R3) and a mouse monoclonal antibromodeoxyuridine antibody to detect newly synthesized RNA.
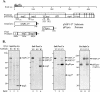
FIG. 4.
Analysis of products expressed from SARS coronavirus pPLpro and pNSP1-3* constructs alone and during coexpression in a _trans_-cleavage assay. (A) Schematic diagram of the predicted processing of SARS coronavirus ORF1a, the constructs expressing the NSP1-3* substrate and PLpro, and the expected final products. (B) Detection of processing by PLpro of the nsp1/2 and nsp2/3 cleavage sites in ORF1a. HeLa-MHVR cells were infected with vTF7.3 and transfected with plasmid DNAs as indicated above each lane. Newly synthesized proteins were labeled with Trans35S label from 5.5 to 10.5 h postinfection. Lysates were prepared and subjected to immunoprecipitation with anti-R1, anti-R2, or anti-V5 antibody. Immunoprecipitated proteins were analyzed by electrophoresis on an SDS-5.0 to 12.5% polyacrylamide gel, processed, and subjected to autoradiography. Products were immunoprecipitated with anti-R1, anti-R2, or anti-V5 epitope tag antibody from cells transfected with DNA encoding the SARS coronavirus NSP1-3* substrate region alone (lanes 1, 4, and 7), the SARS coronavirus PLpro alone (lanes 2, 5, and 8), or cotransfected with both constructs (lanes 3, 6, and 9) or an inactive mutant of PLpro (lane 12). The positions of molecular size markers are shown on the left of each gel (in kilodaltons).
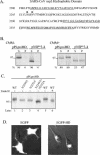
FIG. 5.
Analysis of products expressed from SARS coronavirus pPLpro-HD, pPLpro, and pNSP*3-4 constructs alone and during cotransfection in a _trans_-cleavage assay. (A) Schematic diagram of the predicted processing of SARS coronavirus ORF1a, the constructs expressing PLpro, PLpro-HD, and the NSP*3-4 substrate. (B) Detection of processing by PLpro-HD at the nsp3/4 cleavage site of ORF1a. The _trans_-cleavage assay was performed as described for Fig. 4 and in Materials and Methods. (C) Effect of amino acid substitutions on PLpro activity. Predicted catalytic and control residues (indicated above the lanes) were changed to alanine, plasmid DNA was cotransfected with pNSP*3-4, and products were analyzed by SDS-PAGE (lanes 1 to 4). The effect of changing glycine to alanine in the P1 position of the nsp3/4 cleavage site is shown in lane 6. The positions of molecular size markers are shown on the left of each gel (in kilodaltons).
FIG. 6.
Membrane association of PLpro-HD and NSP*3-4 proteins and N-linked glycosylation of asparagine residues in the hydrophobic domain. (A) Sequence of the SARS coronavirus nsp3 hydrophobic domain (HD, amino acid residues 2207 to 2365). Consensus sites for N-linked glycosylation are indicated by *. Underlined amino acid residues are potential transmembrane sequences (bioinformatics analysis with
). (B) Detection of membrane association of the PLpro-HD and NSP*3-4 substrate. Plasmid DNAs were linearized and subjected to in vitro transcription and translation in the absence or presence of canine microsomal membranes (CMM). Proteins were labeled with Trans-35S label, immunoprecipitated with anti-V5 antibody, analyzed by electrophoresis on SDS-10% polyacrylamide gels, visualized by autoradiography, and quantitated by phosphorimaging. The percentage of total protein detected in the soluble (S) versus the pelleted (P) fraction is indicated below the gel. (C) Effect of amino acid substitution of asparagine residues 2249 and 2252 on the migration of the PLpro-HD protein. Asparagine residues were changed to alanine residues by site-directed mutagenesis as described in Materials and Methods. Wild-type and mutant plasmid DNAs were expressed via the vaccinia virus-T7 expression system, and immunoprecipitated products were analyzed by electrophoresis on SDS-10% polyacrylamide gels. The effect of treatment with endoglycosidase H (Endo H) to remove N-linked glycosylation is shown in lane 6. (D) Detection of EGFP from cells transfected with either pEGFP or pEGFD-HD. HeLa cells were transfected with the indicated DNA, fixed, and visualized by confocal microscopy at 6 h after transfection.
Similar articles
- The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity.
Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. Barretto N, et al. J Virol. 2005 Dec;79(24):15189-98. doi: 10.1128/JVI.79.24.15189-15198.2005. J Virol. 2005. PMID: 16306590 Free PMC article. - Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease.
Yang X, Chen X, Bian G, Tu J, Xing Y, Wang Y, Chen Z. Yang X, et al. J Gen Virol. 2014 Mar;95(Pt 3):614-626. doi: 10.1099/vir.0.059014-0. Epub 2013 Dec 20. J Gen Virol. 2014. PMID: 24362959 - Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63.
Chen Z, Wang Y, Ratia K, Mesecar AD, Wilkinson KD, Baker SC. Chen Z, et al. J Virol. 2007 Jun;81(11):6007-18. doi: 10.1128/JVI.02747-06. Epub 2007 Mar 28. J Virol. 2007. PMID: 17392370 Free PMC article. - The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds.
Báez-Santos YM, St John SE, Mesecar AD. Báez-Santos YM, et al. Antiviral Res. 2015 Mar;115:21-38. doi: 10.1016/j.antiviral.2014.12.015. Epub 2014 Dec 29. Antiviral Res. 2015. PMID: 25554382 Free PMC article. Review. - Spatial and temporal roles of SARS-CoV PLpro -A snapshot.
Yan S, Wu G. Yan S, et al. FASEB J. 2021 Jan;35(1):e21197. doi: 10.1096/fj.202002271. FASEB J. 2021. PMID: 33368679 Free PMC article. Review.
Cited by
- Inhibition of SARS-CoV-2 3CL Mpro by Natural and Synthetic Inhibitors: Potential Implication for Vaccine Production Against COVID-19.
Ullah A, Ullah K. Ullah A, et al. Front Mol Biosci. 2021 Apr 12;8:640819. doi: 10.3389/fmolb.2021.640819. eCollection 2021. Front Mol Biosci. 2021. PMID: 33912587 Free PMC article. - Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV.
Lee H, Lei H, Santarsiero BD, Gatuz JL, Cao S, Rice AJ, Patel K, Szypulinski MZ, Ojeda I, Ghosh AK, Johnson ME. Lee H, et al. ACS Chem Biol. 2015 Jun 19;10(6):1456-65. doi: 10.1021/cb500917m. Epub 2015 Mar 16. ACS Chem Biol. 2015. PMID: 25746232 Free PMC article. - Mutational profiling of SARS-CoV-2 papain-like protease reveals requirements for function, structure, and drug escape.
Wu X, Go M, Nguyen JV, Kuchel NW, Lu BGC, Zeglinski K, Lowes KN, Calleja DJ, Mitchell JP, Lessene G, Komander D, Call ME, Call MJ. Wu X, et al. Nat Commun. 2024 Jul 23;15(1):6219. doi: 10.1038/s41467-024-50566-9. Nat Commun. 2024. PMID: 39043718 Free PMC article. - Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease?
Sulea T, Lindner HA, Purisima EO, Ménard R. Sulea T, et al. J Virol. 2005 Apr;79(7):4550-1. doi: 10.1128/JVI.79.7.4550-4551.2005. J Virol. 2005. PMID: 15767458 Free PMC article. No abstract available. - Deubiquitinating activity of the SARS-CoV papain-like protease.
Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. Barretto N, et al. Adv Exp Med Biol. 2006;581:37-41. doi: 10.1007/978-0-387-33012-9_5. Adv Exp Med Biol. 2006. PMID: 17037501 Free PMC article. No abstract available.
References
- Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. - PubMed
- Bonilla, P. J., S. A. Hughes, J. D. Pinon, and S. R. Weiss. 1995. Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site. Virology 209:489-497. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous