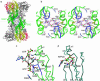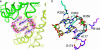Structural basis of activity and allosteric control of diguanylate cyclase - PubMed (original) (raw)
Structural basis of activity and allosteric control of diguanylate cyclase
Carmen Chan et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Recent discoveries suggest that a novel second messenger, bis-(3'-->5')-cyclic di-GMP (c-diGMP), is extensively used by bacteria to control multicellular behavior. Condensation of two GTP to the dinucleotide is catalyzed by the widely distributed diguanylate cyclase (DGC or GGDEF) domain that occurs in various combinations with sensory and/or regulatory modules. The crystal structure of the unorthodox response regulator PleD from Caulobacter crescentus, which consists of two CheY-like receiver domains and a DGC domain, has been solved in complex with the product c-diGMP. PleD forms a dimer with the CheY-like domains (the stem) mediating weak monomer-monomer interactions. The fold of the DGC domain is similar to adenylate cyclase, but the nucleotide-binding mode is substantially different. The guanine base is H-bonded to Asn-335 and Asp-344, whereas the ribosyl and alpha-phosphate moieties extend over the beta2-beta3-hairpin that carries the GGEEF signature motif. In the crystal, c-diGMP molecules are crosslinking active sites of adjacent dimers. It is inferred that, in solution, the two DGC domains of a dimer align in a two-fold symmetric way to catalyze c-diGMP synthesis. Two mutually intercalated c-diGMP molecules are found tightly bound at the stem-DGC interface. This allosteric site explains the observed noncompetitive product inhibition. We propose that product inhibition is due to domain immobilization and sets an upper limit for the concentration of this second messenger in the cell.
Figures
Fig. 1.
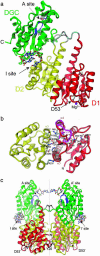
Crystal structure of PleD. (a) The monomer consists of three domains connected by disordered linker peptides (light gray). Domains D1 (residues 2–140, red) and D2 (residues 141–285, yellow) show the CheY-like fold. D1 carries the phosphoacceptor D53. The catalytic DGC domain (286–454) is shown in green. The GGEEF signature motif is located on the β-hairpin (blue) and constitutes part of the active site (A-site) to which a c-diGMP molecule is bound. Two c-diGMP molecules are found at the D2/DGC interface (I-site). (b) The D1(red)/D2(yellow) interface as viewed along the quasi-two-fold axis. Compared with the view in a, the structure has been rotated by 90° approximately around the horizontal. Ionic residues in the interface and residues implicated in activation (phosphoacceptor D53, K105, T83, and F102) are shown. The trace of the β4-α4 loop and F101 of phosphorylated FixJ [magenta; PDB ID code 1d5w (23)] is shown superimposed on D1. (c) The two monomers of the asymmetric unit form a two-fold dimer. The view is related by a –60° rotation about the dimer (vertical) axis with respect to the view in a. The c-diGMP molecules that are bound to sites A and A′ crosslink to another dimer above (see Fig. 2_a_).
Fig. 2.
Ligand binding to the active site of PleD. (a) Dimers are packed head-to-head to form 222 tetramers. The view is rotated by 135° about the vertical axis with respect to Fig. 1_c_. The dimers are held together by two c-diGMP ligands that are located on a local two-fold axis of the tetramer (the viewing direction), the ligand molecule in the back has been omitted for clarity. (b) Stereographic close-up view of a. The G368GEEF signature motif comprises residues important for substrate binding (G368, G369, and E371) and catalysis (E370). The omit map of the ligand is contoured at 4 σ. The side chains of E370, E371, R300, K332, and D336 are partly disordered. (c) Complex of DGC with substrate GTP-Mg as modeled on the basis of the product complex shown in b. The positions of the guanine, ribose, and α-phosphoryl moieties are the same as in the product complex structure. The upper substrate, which has been shifted arbitrarily by ≈2 Å to the upper right, would be bound to another two-fold related DGC domain (not shown). The side-chain conformations of E370 and K332 have been adjusted to bring the functional groups into catalytically competent positions. The arrows indicate the nucleophilic attack of the 3′-oxygens on the α-phosphates. (d) AC in complex with substrate analog ATP-α-S [light-blue, PDB ID code 1cjk (25)], view corresponding to that in c. The Cα-trace corresponds to that part of the α-chain, which is structurally homologous to DGC. The β-chain, which provides specific interactions with the adenine base, has been omitted for clarity.
Fig. 3.
Product binding to the allosteric inhibitory I-site. (a) A close-up view of the two mutually intercalated c-diGMP molecules (khaki and gray carbon atoms) bound at the D2 (yellow)–DGC (green) interface. The omit map of the ligand is contoured at 4 σ.(b) The ligand is tightly bound to both domains [carbons are colored in magenta (D2) and cyan (DGC)] by a multitude of specific interactions, including a recurrent arginine–guanine-binding motif. Figures were generated by
dino
(A. Philippsen,
).
Fig. 4.
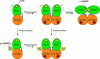
Mechanistic model of PleD regulation. The catalytic DGC domain (green) is tethered via a flexible linker peptide to the D1/D2 stem. The DGC domain is postulated to be mobile with respect to the stem, as indicated by the curved broken arrow. (Upper) PleD is activated by phosphorylation at the D1 domain, which induces dimerization mediated by the stems and allows the two substrate-binding sites (with bound GTP substrate in yellow) to approach each other and the condensation reaction (2 GTP → c-diGMP + 2 PPi) to occur. (Lower) Allosteric product inhibition occurs by binding of (c-diGMP)2 to the I-site at the stem–DGC interface, whereby the DGC domain is immobilized with respect to the stem and barred from approaching its counterpart in the dimer.
Similar articles
- Structure of BeF3- -modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition.
Wassmann P, Chan C, Paul R, Beck A, Heerklotz H, Jenal U, Schirmer T. Wassmann P, et al. Structure. 2007 Aug;15(8):915-27. doi: 10.1016/j.str.2007.06.016. Structure. 2007. PMID: 17697997 - The structure and inhibition of a GGDEF diguanylate cyclase complexed with (c-di-GMP)(2) at the active site.
Yang CY, Chin KH, Chuah ML, Liang ZX, Wang AH, Chou SH. Yang CY, et al. Acta Crystallogr D Biol Crystallogr. 2011 Dec;67(Pt 12):997-1008. doi: 10.1107/S090744491104039X. Epub 2011 Nov 18. Acta Crystallogr D Biol Crystallogr. 2011. PMID: 22120736 - In Silico Discovery and In Vitro Validation of Catechol-Containing Sulfonohydrazide Compounds as Potent Inhibitors of the Diguanylate Cyclase PleD.
Fernicola S, Paiardini A, Giardina G, Rampioni G, Leoni L, Cutruzzolà F, Rinaldo S. Fernicola S, et al. J Bacteriol. 2015 Sep 28;198(1):147-56. doi: 10.1128/JB.00742-15. Print 2016 Jan 1. J Bacteriol. 2015. PMID: 26416830 Free PMC article. - C-di-GMP Synthesis: Structural Aspects of Evolution, Catalysis and Regulation.
Schirmer T. Schirmer T. J Mol Biol. 2016 Sep 25;428(19):3683-701. doi: 10.1016/j.jmb.2016.07.023. Epub 2016 Aug 4. J Mol Biol. 2016. PMID: 27498163 Review. - Structures, mechanism, regulation and evolution of class III nucleotidyl cyclases.
Sinha SC, Sprang SR. Sinha SC, et al. Rev Physiol Biochem Pharmacol. 2006;157:105-40. doi: 10.1007/112_0603. Rev Physiol Biochem Pharmacol. 2006. PMID: 17236651 Review.
Cited by
- Control of Biofilm Formation by an Agrobacterium tumefaciens Pterin-Binding Periplasmic Protein Conserved Among Pathogenic Bacteria.
Greenwich JL, Eagan JL, Feirer N, Boswinkle K, Minasov G, Shuvalova L, Inniss NL, Raghavaiah J, Ghosh AK, Satchell KJF, Allen KD, Fuqua C. Greenwich JL, et al. bioRxiv [Preprint]. 2023 Nov 21:2023.11.18.567607. doi: 10.1101/2023.11.18.567607. bioRxiv. 2023. PMID: 38014264 Free PMC article. Updated. Preprint. - Systematic Nomenclature for GGDEF and EAL Domain-Containing Cyclic Di-GMP Turnover Proteins of Escherichia coli.
Hengge R, Galperin MY, Ghigo JM, Gomelsky M, Green J, Hughes KT, Jenal U, Landini P. Hengge R, et al. J Bacteriol. 2015 Jul 6;198(1):7-11. doi: 10.1128/JB.00424-15. Print 2016 Jan 1. J Bacteriol. 2015. PMID: 26148715 Free PMC article. - The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control.
Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. Lindenberg S, et al. EMBO J. 2013 Jul 17;32(14):2001-14. doi: 10.1038/emboj.2013.120. Epub 2013 May 24. EMBO J. 2013. PMID: 23708798 Free PMC article. - Structural basis of ligand binding by a c-di-GMP riboswitch.
Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Smith KD, et al. Nat Struct Mol Biol. 2009 Dec;16(12):1218-23. doi: 10.1038/nsmb.1702. Epub 2009 Nov 8. Nat Struct Mol Biol. 2009. PMID: 19898477 Free PMC article. - Adaptive divergence in experimental populations of Pseudomonas fluorescens. II. Role of the GGDEF regulator WspR in evolution and development of the wrinkly spreader phenotype.
Goymer P, Kahn SG, Malone JG, Gehrig SM, Spiers AJ, Rainey PB. Goymer P, et al. Genetics. 2006 Jun;173(2):515-26. doi: 10.1534/genetics.106.055863. Epub 2006 Apr 19. Genetics. 2006. PMID: 16624907 Free PMC article.
References
- Uzzau, S. & Fasano, A. (2000) Cell Microbiol. 2, 83–89. - PubMed
- Ross, P., Weinhouse, H., Aloni, Y., Michaeli, D., Weinberger-Ohana, P., Mayer, R., Braun, S., De Vroom, E., Van der Marel, G. A., Van Boom, J. H., et al. (1987) Nature 325, 279–281. - PubMed
- Jenal, U. (2004) Curr. Opin. Microbiol. 7, 185–191. - PubMed
- Simm, R., Morr, M., Kader, A., Nimtz, M. & Romling, U. (2004) Mol. Microbiol. 53, 1123–1134. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous