Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome - PubMed (original) (raw)
Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome
Brian P Chadwick et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Heterochromatin is defined classically by condensation throughout the cell cycle, replication in late S phase and gene inactivity. Facultative heterochromatin is of particular interest, because its formation is developmentally regulated as a result of cellular differentiation. The most extensive example of facultative heterochromatin is the mammalian inactive X chromosome (Xi). A variety of histone variants and covalent histone modifications have been implicated in defining the organization of the Xi heterochromatic state, and the features of Xi heterochromatin have been widely interpreted as reflecting a redundant system of gene silencing. However, here we demonstrate that the human Xi is packaged into at least two nonoverlapping heterochromatin types, each characterized by specific Xi features: one defined by the presence of Xi-specific transcript RNA, the histone variant macroH2A, and histone H3 trimethylated at lysine 27 and the other defined by H3 trimethylated at lysine 9, heterochromatin protein 1, and histone H4 trimethylated at lysine 20. Furthermore, regions of the Xi packaged in different heterochromatin types are characterized by different patterns of replication in late S phase. The arrangement of facultative heterochromatin into spatially and temporally distinct domains has implications for both the establishment and maintenance of the Xi and adds a previously unsuspected degree of epigenetic complexity.
Figures
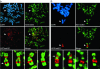
Fig. 1.
Spatial relationship of two major Xi heterochromatin types at metaphase. Images represent typical distributions obtained from three independent female cell lines. (a) Partial metaphase spread of RPE1 cells showing the spatial distribution of H3TrimK9 (green, FITC) and H3TrimK27 (red, rhodamine) and four additional higher-magnification images of the Xi showing the merged H3TrimK9 and H3TrimK27 distributions. The white arrow indicates the major H3TrimK27 band centered at Xq23. (b) Distributions of H3TrimK9 and H3TrimK27 in HME1 cells. The location of the Xi in the partial metaphase spreads is indicated by white arrowheads. The white arrow indicates the major H3TrimK27 band centered at Xp11. All images were obtained by indirect immunofluorescence.
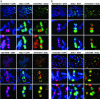
Fig. 2.
Correlation of H3TrimK27 (a Upper and b Upper) and H3TrimK9 (a Lower and b Lower) heterochromatin with the Xi replication pattern. (a) Metaphase chromosomes prepared from RPE1 cells that were incubated with BrdUrd for 4 h before metaphase arrest (corresponding approximately to the last 2 h of S phase). (b) Metaphase chromosomes prepared from HME1 cells that were incubated with BrdUrd for 3 h before metaphase arrest (corresponding approximately to the last 2 h of S phase). In each section, Top shows a partial metaphase spread, whereas Middle and Bottom show the Xi from independent spreads at higher magnification.
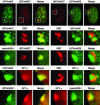
Fig. 3.
Characterization of Xi chromatin territories at interphase. All images are of the RPE1 cells and represent typical observations made in at least four independent female cell lines. The white box in each interphase nucleus (top row) represents the Barr body region examined at higher magnification in the other images below. The feature examined by indirect immunofluorescence is labeled above each image. Overlapping red and green signals appear yellow.
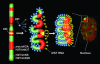
Fig. 4.
Schematic model showing how heterochromatin of the Xi could transition between metaphase and interphase to be organized into the two nonoverlapping heterochromatin territories and to explain how XIST RNA could rapidly spread in cis outward from the X inactivation center (XIC) along only part of the Xi. See main text for details.
Similar articles
- Chromatin of the Barr body: histone and non-histone proteins associated with or excluded from the inactive X chromosome.
Chadwick BP, Willard HF. Chadwick BP, et al. Hum Mol Genet. 2003 Sep 1;12(17):2167-78. doi: 10.1093/hmg/ddg229. Epub 2003 Jul 15. Hum Mol Genet. 2003. PMID: 12915472 - Histone acetylation controls the inactive X chromosome replication dynamics.
Casas-Delucchi CS, Brero A, Rahn HP, Solovei I, Wutz A, Cremer T, Leonhardt H, Cardoso MC. Casas-Delucchi CS, et al. Nat Commun. 2011;2:222. doi: 10.1038/ncomms1218. Nat Commun. 2011. PMID: 21364561 Free PMC article. - Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin.
Peters AH, Mermoud JE, O'Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T. Peters AH, et al. Nat Genet. 2002 Jan;30(1):77-80. doi: 10.1038/ng789. Epub 2001 Dec 10. Nat Genet. 2002. PMID: 11740497 - Histone modifications and nuclear architecture: a review.
Bártová E, Krejcí J, Harnicarová A, Galiová G, Kozubek S. Bártová E, et al. J Histochem Cytochem. 2008 Aug;56(8):711-21. doi: 10.1369/jhc.2008.951251. Epub 2008 May 12. J Histochem Cytochem. 2008. PMID: 18474937 Free PMC article. Review. - X chromosome inactivation: heterogeneity of heterochromatin.
Sidhu SK, Minks J, Chang SC, Cotton AM, Brown CJ. Sidhu SK, et al. Biochem Cell Biol. 2008 Oct;86(5):370-9. doi: 10.1139/o08-100. Biochem Cell Biol. 2008. PMID: 18923538 Review.
Cited by
- Control of gene expression and assembly of chromosomal subdomains by chromatin regulators with antagonistic functions.
Lam AL, Pazin DE, Sullivan BA. Lam AL, et al. Chromosoma. 2005 Sep;114(4):242-51. doi: 10.1007/s00412-005-0001-0. Epub 2005 Oct 15. Chromosoma. 2005. PMID: 16012860 Review. - Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome.
Sarma K, Levasseur P, Aristarkhov A, Lee JT. Sarma K, et al. Proc Natl Acad Sci U S A. 2010 Dec 21;107(51):22196-201. doi: 10.1073/pnas.1009785107. Epub 2010 Dec 6. Proc Natl Acad Sci U S A. 2010. PMID: 21135235 Free PMC article. - Diverse factors are involved in maintaining X chromosome inactivation.
Chan KM, Zhang H, Malureanu L, van Deursen J, Zhang Z. Chan KM, et al. Proc Natl Acad Sci U S A. 2011 Oct 4;108(40):16699-704. doi: 10.1073/pnas.1107616108. Epub 2011 Sep 21. Proc Natl Acad Sci U S A. 2011. PMID: 21940502 Free PMC article. - Emerging Roles of Repetitive and Repeat-Containing RNA in Nuclear and Chromatin Organization and Gene Expression.
Trigiante G, Blanes Ruiz N, Cerase A. Trigiante G, et al. Front Cell Dev Biol. 2021 Oct 6;9:735527. doi: 10.3389/fcell.2021.735527. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34722514 Free PMC article. Review. - Histone H3 trimethylation at lysine 36 is associated with constitutive and facultative heterochromatin.
Chantalat S, Depaux A, Héry P, Barral S, Thuret JY, Dimitrov S, Gérard M. Chantalat S, et al. Genome Res. 2011 Sep;21(9):1426-37. doi: 10.1101/gr.118091.110. Epub 2011 Jul 29. Genome Res. 2011. PMID: 21803857 Free PMC article.
References
- Lyon, M. F. (1961) Nature 190, 372–373. - PubMed
- Avner, P. & Heard, E. (2001) Nat. Rev. 2, 59–67. - PubMed
- Plath, K., Mlynarczyk-Evans, S., Nusinow, D. A. & Panning, B. (2002) Annu. Rev. Genet. 36, 233–278. - PubMed
- Chadwick, B. P. & Willard, H. F. (2001) Hum. Mol. Genet. 10, 1101–1013. - PubMed
- Costanzi, C. & Pehrson, J. R. (1998) Nature 393, 599–601. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials