Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila - PubMed (original) (raw)
Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila
Anja Ebert et al. Genes Dev. 2004.
Abstract
Histone lysine methylation is an epigenetic mark to index chromosomal subdomains. In Drosophila, H3-K9 di- and trimethylation is mainly controlled by the heterochromatic SU(VAR)3-9 HMTase, a major regulator of position-effect variegation (PEV). In contrast, H3-K27 methylation states are independently mediated by the Pc-group enzyme E(Z). Isolation of 19 point mutants demonstrates that the silencing potential of Su(var)3-9 increases with its associated HMTase activity. A hyperactive Su(var)3-9 mutant, pitkin(D), displays extensive H3-K9 di- and trimethylation within but also outside pericentric heterochromatin. Notably, mutations in a novel Su(var) gene, Su(var)3-1, severely restrict Su(var)3-9-mediated gene silencing. Su(var)3-1 was identified as "antimorphic" mutants of the euchromatic H3-S10 kinase JIL-1. JIL-1(Su(var)3-1) mutants maintain kinase activity and do not detectably impair repressive histone lysine methylation marks. However, analyses with seven different PEV rearrangements demonstrate a general role of JIL-1(Su(var)3-1) in controlling heterochromatin compaction and expansion. Our data provide evidence for a dynamic balance between heterochromatin and euchromatin, and define two distinct mechanisms for Su(var) gene function. Whereas the majority of Su(var)s encode inherent components of heterochromatin that can establish repressive chromatin structures [intrinsic Su(var)s], Su(var)3-1 reflects gain-of-function mutants of a euchromatic component that antagonize the expansion of heterochromatic subdomains [acquired Su(var)s].
Figures
Figure 1.
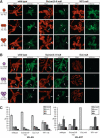
Control of H3-K9 and H3-K27 methylation states in Drosophila heterochromatin. (A) Distribution of H3-K9 mono-, di-, and trimethylation in the chromocenter region of salivary gland polytene chromosomes from wild-type, _Su(var)3-9_-null, and HP1-null third instar larvae. (B) Distribution of H3-K27 mono-, di-, and trimethylation in the chromocenter region of polytene chromosomes from wild-type and _Su(var)3-9_-null as well as in whole chromosomes of _E(z)_-null third instar larvae. DNA was stained with propidium iodide. Arrows point to the fourth chromosome; arrowheads indicate the chromocenter core region. (C) Comparative mass spectometric analyses of H3-K9 and H3-K27 mono-, di-, and trimethylation levels in H3 peptide fragments isolated from nuclear extracts of wild-type, _Su(var)3-9_-null, _E(z)_-null, and HP1-null mutant larvae. For each genotype three different extracts were analyzed (error bar indicates standard deviation). The relative level of a particular H3-K9 methylation state is indicated with respect to the sum of all H3-K9 and H3-K27 modifications present in the H3 peptide populations.
Figure 2.
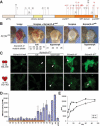
SU(VAR)3-9 HMTase activity correlates with its ability to induce gene silencing. (A) Protein structure of SU(VAR)3-9 and positions of the molecularly characterized mutations. Point mutations resulting in amino acid exchanges are in red. (B) Genetic classification of Su(var)3-9 mutants. Su(var)3-9mut alleles were crossed to two extra Su(var)3-9 copies. Null mutants show the characteristic white mottled phenotype, whereas hypomorphic alleles partially enhance white repression. Su(var)3-9ptn is a hypermorphic mutant resulting in complete white and partial silencing of the roughest gene. (C) H3-K9 di- and trimethylation in chromocenter regions of heterozygous Su(var)3-917/Su(var)3-9mut mutants. Null mutants [exemplified by _Su(var)3-910_] show the same loss of H3-K9 di- and trimethylation as the _Su(var)3-917/Su(var)3-917_-null mutant. H3-K9 methylation levels are elevated in hypomorphic mutants [exemplified by _Su(var)3-922_]. Very high levels of H3-K9 di- and trimethylation are found in hypermorphic Su(var)3-9ptn. Arrowheads indicate the chromocenter core. (D) In vitro HMTase activity of recombinant SU(VAR)3-9 mutant proteins. The amount of incorporated label was measured by scintillation counting. (E) Time-course analysis of HMTase reactions with wild-type (wt) and SU(VAR)3-9ptn proteins. Incorporation of the radioactive label was measured after different timepoints by scintillation counting.
Figure 3.
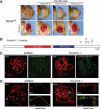
Su(var)3-1 mutants are antimorphic alleles of the JIL-1 kinase. (A) Genomic extra copies of Su(var)3-9, Su(var)2-5, and Su(var)3-7 display enhanced silencing of the white gene in In(1)wm4 (upper row), which is impaired in the presence of the strong PEV suppressor mutant Su(var)3-1 (lower row). (B) Protein structure of JIL-1. All mutational lesions in Su(var)3-1 alleles result in a C-terminally truncated JIL-1 protein. (S/T) Serine/threonine kinase domain; (S/T/Y) Serine/threonine/tyrosine kinase domain. (C) Polytene chromosomes of wild-type and Su(var)3-1 mutant larvae were stained with pH3S10-specific antibodies. DNA was stained with propidium iodide. (D) X-chromosome decondensation phenotype of Su(var)3-1 mutants. In male homozygous mutant larvae, the X-chromosome is largely decondensed and partially loses its banding structure. The arrowhead points to the X-chromosome. The affected chromosome shows association with MOF, a specific marker for the male X-chromosome. DNA was stained with propidium iodide. Bar, 10 μm.
Figure 4.
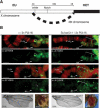
Su(var)3-1 mutants impair expansion of heterochromatin. (A) Translocation T(1;4)wm258-21 contains the X-chromosomal region 3C-3E comprising ∼1 Mb of DNA, which can be detected because of contacts to the fourth chromosome. Heterochromatin expansion inactivates the two marker genes Notch (clipped wings) and white (white eyes). (B) Immunocytological analysis of H3-K9 di- and trimethylation at T(1;4)wm258-21. (Left panel) In the presence of two extra Su(var)3-9 copies, region 3C-3E becomes compacted (bracket) and marked with H3-K9 dimethylation but not H3-K9 trimethylation. Transcriptional repression is indicated by a large number of Notch wings (51%) and white mottled sectored eyes (41%). (Right panel) The presence of Su(var)3-1 mutations impairs expansion of heterochromatin (bracket), resulting in a normal banding pattern, loss of H3-K9 dimethylation, and restored transcriptional activity as indicated by wild-type (Notch+) wings (91%) and red (white+) eyes (95%).
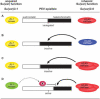
Figure 5.
Su(var) genes regulate the balance between euchromatin and heterochromatin. The antagonistic functions of acquired and intrinsic Su(var) genes establish a boundary between euchromatin and heterochromatin. Hyperactive SU(VAR)3-9ptn (B) or overexpression of Su(var)3-9 (C) can expand heterochromatin, whereas JIL-1Su(var)3-1 stabilizes the euchromatic state by acting on an unknown target protein X (D). Please see main text for details.
Similar articles
- Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3.
Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schäfer C, Phalke S, Walther M, Schmidt A, Jenuwein T, Reuter G. Rudolph T, et al. Mol Cell. 2007 Apr 13;26(1):103-15. doi: 10.1016/j.molcel.2007.02.025. Mol Cell. 2007. PMID: 17434130 - Histone modification and the control of heterochromatic gene silencing in Drosophila.
Ebert A, Lein S, Schotta G, Reuter G. Ebert A, et al. Chromosome Res. 2006;14(4):377-92. doi: 10.1007/s10577-006-1066-1. Chromosome Res. 2006. PMID: 16821134 Review. - Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing.
Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Schotta G, et al. EMBO J. 2002 Mar 1;21(5):1121-31. doi: 10.1093/emboj/21.5.1121. EMBO J. 2002. PMID: 11867540 Free PMC article. - Role of Drosophila HP1 in euchromatic gene expression.
Cryderman DE, Grade SK, Li Y, Fanti L, Pimpinelli S, Wallrath LL. Cryderman DE, et al. Dev Dyn. 2005 Mar;232(3):767-74. doi: 10.1002/dvdy.20310. Dev Dyn. 2005. PMID: 15704177 - SU(VAR)3-9 is a conserved key function in heterochromatic gene silencing.
Schotta G, Ebert A, Reuter G. Schotta G, et al. Genetica. 2003 Mar;117(2-3):149-58. doi: 10.1023/a:1022923508198. Genetica. 2003. PMID: 12723694 Review.
Cited by
- Age-related changes in polycomb gene regulation disrupt lineage fidelity in intestinal stem cells.
Tauc HM, Rodriguez-Fernandez IA, Hackney JA, Pawlak M, Ronnen Oron T, Korzelius J, Moussa HF, Chaudhuri S, Modrusan Z, Edgar BA, Jasper H. Tauc HM, et al. Elife. 2021 Mar 16;10:e62250. doi: 10.7554/eLife.62250. Elife. 2021. PMID: 33724181 Free PMC article. - Enrichment of HP1a on Drosophila chromosome 4 genes creates an alternate chromatin structure critical for regulation in this heterochromatic domain.
Riddle NC, Jung YL, Gu T, Alekseyenko AA, Asker D, Gui H, Kharchenko PV, Minoda A, Plachetka A, Schwartz YB, Tolstorukov MY, Kuroda MI, Pirrotta V, Karpen GH, Park PJ, Elgin SC. Riddle NC, et al. PLoS Genet. 2012 Sep;8(9):e1002954. doi: 10.1371/journal.pgen.1002954. Epub 2012 Sep 20. PLoS Genet. 2012. PMID: 23028361 Free PMC article. - Loss-of-function alleles of the JIL-1 kinase are strong suppressors of position effect variegation of the wm4 allele in Drosophila.
Lerach S, Zhang W, Bao X, Deng H, Girton J, Johansen J, Johansen KM. Lerach S, et al. Genetics. 2006 Aug;173(4):2403-6. doi: 10.1534/genetics.106.059253. Epub 2006 May 15. Genetics. 2006. PMID: 16702418 Free PMC article. Review. - Control of gene expression and assembly of chromosomal subdomains by chromatin regulators with antagonistic functions.
Lam AL, Pazin DE, Sullivan BA. Lam AL, et al. Chromosoma. 2005 Sep;114(4):242-51. doi: 10.1007/s00412-005-0001-0. Epub 2005 Oct 15. Chromosoma. 2005. PMID: 16012860 Review. - The profile of repeat-associated histone lysine methylation states in the mouse epigenome.
Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. Martens JH, et al. EMBO J. 2005 Feb 23;24(4):800-12. doi: 10.1038/sj.emboj.7600545. Epub 2005 Jan 27. EMBO J. 2005. PMID: 15678104 Free PMC article.
References
- Aagaard L., Schmid, M., Warburton, P., and Jenuwein, T. 2000. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J. Cell Sci. 113: 817-829. - PubMed
- Allshire R.C., Javerzat, J.P., Redhead, N.J., and Cranston, G. 1994. Position effect variegation at fission yeast centromeres. Cell 76: 157-169. - PubMed
- Carrington E.A. and Jones, R.S. 1996. The Drosophila Enhancer of zeste gene encodes a chromosomal protein: Examination of wild-type and mutant protein distribution. Development 122: 4073-4083. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases