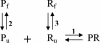Mechanistic approach to the problem of hybridization efficiency in fluorescent in situ hybridization - PubMed (original) (raw)
Mechanistic approach to the problem of hybridization efficiency in fluorescent in situ hybridization
L Safak Yilmaz et al. Appl Environ Microbiol. 2004 Dec.
Abstract
In fluorescent in situ hybridization (FISH), the efficiency of hybridization between the DNA probe and the rRNA has been related to the accessibility of the rRNA when ribosome content and cell permeability are not limiting. Published rRNA accessibility maps show that probe brightness is sensitive to the organism being hybridized and the exact location of the target site and, hence, it is highly unpredictable based on accessibility only. In this study, a model of FISH based on the thermodynamics of nucleic acid hybridization was developed. The model provides a mechanistic approach to calculate the affinity of the probe to the target site, which is defined as the overall Gibbs free energy change (DeltaG degrees overall) for a reaction scheme involving the DNA-rRNA and intramolecular DNA and rRNA interactions that take place during FISH. Probe data sets for the published accessibility maps and experiments targeting localized regions in the 16S rRNA of Escherichia coli were used to demonstrate that DeltaG degrees overall is a strong predictor of hybridization efficiency and superior to conventional estimates based on the dissociation temperature of the DNA/rRNA duplex. The use of the proposed model also allowed the development of mechanistic approaches to increase probe brightness, even in seemingly inaccessible regions of the 16S rRNA. Finally, a threshold DeltaG degrees overall of -13.0 kcal/mol was proposed as a goal in the design of FISH probes to maximize hybridization efficiency without compromising specificity.
Figures
FIG. 1.
Proposed reaction scheme for FISH. P, R, and PR denote the DNA probe, the target (rRNA), and the DNA/rRNA hybrid, respectively. The subscripts f and u represent folded and unfolded conformations.
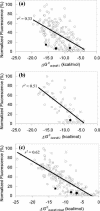
FIG. 2.
Relation between normalized fluorescence intensity of fluorescein-labeled probes (13) and Δ_G_°overall. (a) Complete set of 176 probes; (b) probes targeting sites relatively free of protein contact; (c) all 176 probes when Δ_G_°3 estimates were mixed to maximize r2. The solid diamond, circle, and square symbols denote probes Eco 1014, Eco 1184, and Eco 1274, respectively.
FIG. 3.
Fluorescence intensity as a function of probe affinity for probes from Fuchs et al. (13) targeting helix 6 (open circles, solid regression line) and helix 18 (solid squares, dashed regression line). The dotted curve represents equation 9 fitted to the data by assuming that a [PR_]/_[_R0_] ratio of 1 corresponds to 75% fluorescence and a [PR_]/_[_R0_] ratio of 0 represents 3% fluorescence in Fuchs et al.'s normalized scale.
FIG. 4.
Probe brightness as a function of probe affinity in a subregion of helix 9. The dotted curve represents equation 9 fitted to the data by assuming that [PR_]/_[_R0_] values of 1 and 0 corresponded to 1.05 and 0.1 brightness units, respectively.
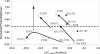
FIG. 5.
Effect of increasing the magnitude of Δ_G_°overall-i on the fluorescence intensity of probes targeting sites located in helix 9 (circles), helices 37 and 38 (squares), helices 39, 43,and 45 (triangles), and helix 46 (diamonds). The data points (except solid circles) are from Table 1. Solid arrows indicate the trend for each target site. The dotted arrow shows the effect of using 30% formamide in the hybridization of E1177. Data points represented by solid circles are from Fuchs et al. (13), and the dashed line shows the equivalency to the lower limit of class 3 in Fuchs et al.'s scale.
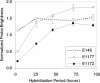
FIG. 6.
Effect of hybridization time on brightness of probes targeting helices 39, 43, and 45. Data points show averages and standard deviations of duplicate hybridizations. Probe E146 was used as a control.
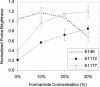
FIG. 7.
Effect of formamide concentration on brightness of probes targeting helices 39, 43, and 45. Data points show averages and standard deviations of two independent experiments. Probe E146 was used as a control.
FIG. 8.
Venn diagram representation of the relation among the factors affecting fluorescence intensity in FISH.
Similar articles
- Making all parts of the 16S rRNA of Escherichia coli accessible in situ to single DNA oligonucleotides.
Yilmaz LS, Okten HE, Noguera DR. Yilmaz LS, et al. Appl Environ Microbiol. 2006 Jan;72(1):733-44. doi: 10.1128/AEM.72.1.733-744.2006. Appl Environ Microbiol. 2006. PMID: 16391113 Free PMC article. - Development of thermodynamic models for simulating probe dissociation profiles in fluorescence in situ hybridization.
Yilmaz LS, Noguera DR. Yilmaz LS, et al. Biotechnol Bioeng. 2007 Feb 1;96(2):349-63. doi: 10.1002/bit.21114. Biotechnol Bioeng. 2007. PMID: 16878331 - Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes.
Fuchs BM, Wallner G, Beisker W, Schwippl I, Ludwig W, Amann R. Fuchs BM, et al. Appl Environ Microbiol. 1998 Dec;64(12):4973-82. doi: 10.1128/AEM.64.12.4973-4982.1998. Appl Environ Microbiol. 1998. PMID: 9835591 Free PMC article. - Prediction of melting temperatures in fluorescence in situ hybridization (FISH) procedures using thermodynamic models.
Fontenete S, Guimarães N, Wengel J, Azevedo NF. Fontenete S, et al. Crit Rev Biotechnol. 2016;36(3):566-77. doi: 10.3109/07388551.2014.993589. Epub 2015 Jan 14. Crit Rev Biotechnol. 2016. PMID: 25586037 Review. - A comprehensive model for the diffusion and hybridization processes of nucleic acid probes in fluorescence in situ hybridization.
Lima JF, Maia P, T Magalhães B, Cerqueira L, Azevedo NF. Lima JF, et al. Biotechnol Bioeng. 2020 Oct;117(10):3212-3223. doi: 10.1002/bit.27462. Epub 2020 Jul 2. Biotechnol Bioeng. 2020. PMID: 32946120 Review.
Cited by
- Spectral imaging and nucleic acid mimics fluorescence in situ hybridization (SI-NAM-FISH) for multiplex detection of clinical pathogens.
Azevedo AS, Fernandes RM, Faria AR, Silvestre OF, Nieder JB, Lou C, Wengel J, Almeida C, Azevedo NF. Azevedo AS, et al. Front Microbiol. 2022 Sep 29;13:976639. doi: 10.3389/fmicb.2022.976639. eCollection 2022. Front Microbiol. 2022. PMID: 36246234 Free PMC article. - probeBase--an online resource for rRNA-targeted oligonucleotide probes: new features 2007.
Loy A, Maixner F, Wagner M, Horn M. Loy A, et al. Nucleic Acids Res. 2007 Jan;35(Database issue):D800-4. doi: 10.1093/nar/gkl856. Epub 2006 Nov 11. Nucleic Acids Res. 2007. PMID: 17099228 Free PMC article. - Detection of tmRNA molecules on microarrays at low temperatures using helper oligonucleotides.
Kaplinski L, Scheler O, Parkel S, Palta P, Toome K, Kurg A, Remm M. Kaplinski L, et al. BMC Biotechnol. 2010 Apr 28;10:34. doi: 10.1186/1472-6750-10-34. BMC Biotechnol. 2010. PMID: 20426847 Free PMC article. - Comparison of vertical distributions of prokaryotic assemblages in the anoxic Cariaco Basin and Black Sea by use of fluorescence in situ hybridization.
Lin X, Wakeham SG, Putnam IF, Astor YM, Scranton MI, Chistoserdov AY, Taylor GT. Lin X, et al. Appl Environ Microbiol. 2006 Apr;72(4):2679-90. doi: 10.1128/AEM.72.4.2679-2690.2006. Appl Environ Microbiol. 2006. PMID: 16597973 Free PMC article. - Characterization of the community structure of a dechlorinating mixed culture and comparisons of gene expression in planktonic and biofloc-associated "Dehalococcoides" and Methanospirillum species.
Rowe AR, Lazar BJ, Morris RM, Richardson RE. Rowe AR, et al. Appl Environ Microbiol. 2008 Nov;74(21):6709-19. doi: 10.1128/AEM.00445-08. Epub 2008 Sep 5. Appl Environ Microbiol. 2008. PMID: 18776027 Free PMC article.
References
- Amann, R., B. M. Fuchs, and S. Behrens. 2001. The identification of microorganisms by fluorescence in situ hybridisation. Curr. Opin. Biotechnol. 12:231-236. - PubMed
- Ban, N., P. Nissen, J. Hansen, P.-B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905-920. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources