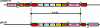Complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity - PubMed (original) (raw)
Complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity
Glenn Rhodes et al. Appl Environ Microbiol. 2004 Dec.
Abstract
This study presents the first complete sequence of an IncU plasmid, pFBAOT6. This plasmid was originally isolated from a strain of Aeromonas caviae from hospital effluent (Westmorland General Hospital, Kendal, United Kingdom) in September 1997 (G. Rhodes, G. Huys, J. Swings, P. McGann, M. Hiney, P. Smith, and R. W. Pickup, Appl. Environ. Microbiol. 66:3883-3890, 2000) and belongs to a group of related plasmids with global ubiquity. pFBAOT6 is 84,748 bp long and has 94 predicted coding sequences, only 12 of which do not have a possible function that has been attributed. Putative replication, maintenance, and transfer functions have been identified and are located in a region in the first 31 kb of the plasmid. The replication region is poorly understood but exhibits some identity at the protein level with replication proteins from the gram-positive bacteria Bacillus and Clostridium. The mating pair formation system is a virB homologue, type IV secretory pathway that is similar in its structural organization to the mating pair formation systems of the related broad-host-range (BHR) environmental plasmids pIPO2, pXF51, and pSB102 from plant-associated bacteria. Partitioning and maintenance genes are homologues of genes in IncP plasmids. The DNA transfer genes and the putative oriT site also exhibit high levels of similarity with those of plasmids pIPO2, pXF51, and pSB102. The genetic load region encompasses 54 kb, comprises the resistance genes, and includes a class I integron, an IS630 relative, and other transposable elements in a 43-kb region that may be a novel Tn1721-flanked composite transposon. This region also contains 24 genes that exhibit the highest levels of identity to chromosomal genes of several plant-associated bacteria. The features of the backbone of pFBAOT6 that are shared with this newly defined group of environmental BHR plasmids suggest that pFBAOT6 may be a relative of this group, but a relative that was isolated from a clinical bacterial environment rather than a plant-associated bacterial environment.
Figures
FIG. 1.
Structural organization of the 84,749-bp plasmid pFBAOT6. Plus sense strand-encoded CDSs are indicated above the plasmid line, while CDSs encoded on the complementary strand are indicated below the plasmid line. The CDS numbers for pFBAOT6 are shown in Table 1. The colors indicate the following: red, DNA metabolism (replication, recombination, and transfer); dark green, membrane and surface associated; yellow, miscellaneous metabolism; orange, conserved hypothetical; light green, unknown; pink, mobile elements; white, antibiotic and antimicrobial resistance; and blue, regulation. Inverted repeats are indicated, as are the various transposable elements with which they are associated. The 5-bp direct repeat (DR) flanking the 43-kb Tn_1721_-based transposon is indicated in red, as are other IRs associated with Tn_1721_. The proposed oriT region is indicated by an open triangle below the sequence.
FIG. 2.
Multiple-sequence alignment of pFBAOT6 replication protein with its closest relatives from Bacillus sp. strain KSM-KP43 (KP-43), plasmid pE88, C. tetani, and plasmid pIPO4. A black background indicates amino acids that are fully conserved, while a gray background indicates amino acids that are 50% conserved. The bar above the sequences indicates the helix-turn-helix motif for ReppFBAOT6.
FIG. 3.
Multiple-sequence alignment of KorBpFBAOT6 with its closest relatives from plasmids pIPO2 and pSB102 and the IncP representative R751. A black background indicates amino acids that are fully conserved, while a gray background indicates amino acids that are 50% conserved. The black and gray bars below the sequence of R751 indicate the helix-turn-helix motif and the IncC interaction domain, respectively. The repeating 18- or 19-amino-acid sequence in KorBpFBAOT6 is enclosed in a box and is labeled I to VI.
FIG. 4.
Proposed oriT region of pFBAOT6 aligned with oriT regions from pXF51, pIPO2, and pSB102. The nucleotide numbers for the region in pFBAOT6 are indicated on the left. A black background indicates amino acids that are fully conserved, while a gray background indicates amino acids that are 50% conserved. Inverted repeat sequences are shown only for pFBAOT6 and are indicated by arrows above the sequence and numbers corresponding to the repeats. The nic site is indicated by a solid triangle above the sequence.
FIG. 5.
Translated nucleotide sequence of the IS_630_ family transposon found in pFBAOT6. Inverted repeat ends are indicated by italics. The underlined region is the H-T-H motif. The conserved residues of the DDE motif of the transposase are indicated by larger type.
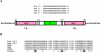
FIG. 6.
Comparison of class I integrons contained in pFBAOT6 and the related IncU plasmid pRAS1. The In_4_-like region shared by the two plasmids is almost identical except for the presence of the dfrA16 cassette in pRAS1 instead of the aadA2 gene in pFBAOT6. The integron in pFBAOT6 is flanked by complete res genes (having separate origins), and res is identical over the shared length to the truncated resA gene of pRAS1.
FIG. 7.
Organization of the composite transposon that is inserted into Tn_1721_-L. (A) The composite transposon is flanked by IS_4_ family IS elements (ISL and ISR) that contain the inverted repeats IR1 to IR4. The green region represents CDS pFBAOT6.52. (B) Regions of the transposase proteins of ISL and ISR aligned with the regions that exhibit the highest levels of identity, highlighting the DDE domain. The numbers in parentheses indicate the numbers of amino acids between the two conserved D residues and between the second conserved D residue and the conserved E residue (circled in the consensus [Consens.] sequence). IS_SB1_ is from plasmid p37 in marine psychrophilic bacterium Mst37 (accession number AJ305328); IS_H8_ and IS_H8A_ are found in Halobacterium sp. strain NRC-1 plasmid pNRC100 (accession number AF016485).
Similar articles
- The complete nucleotide sequence and environmental distribution of the cryptic, conjugative, broad-host-range plasmid pIPO2 isolated from bacteria of the wheat rhizosphere.
Tauch A, Schneiker S, Selbitschka W, Pühler A, van Overbeek LS, Smalla K, Thomas CM, Bailey MJ, Forney LJ, Weightman A, Ceglowski P, Pembroke T, Tietze E, Schröder G, Lanka E, van Elsas JD. Tauch A, et al. Microbiology (Reading). 2002 Jun;148(Pt 6):1637-1653. doi: 10.1099/00221287-148-6-1637. Microbiology (Reading). 2002. PMID: 12055285 - Sequence of the 68,869 bp IncP-1alpha plasmid pTB11 from a waste-water treatment plant reveals a highly conserved backbone, a Tn402-like integron and other transposable elements.
Tennstedt T, Szczepanowski R, Krahn I, Pühler A, Schlüter A. Tennstedt T, et al. Plasmid. 2005 May;53(3):218-38. doi: 10.1016/j.plasmid.2004.09.004. Epub 2004 Dec 9. Plasmid. 2005. PMID: 15848226 - Plasmid pB8 is closely related to the prototype IncP-1beta plasmid R751 but transfers poorly to Escherichia coli and carries a new transposon encoding a small multidrug resistance efflux protein.
Schlüter A, Heuer H, Szczepanowski R, Poler SM, Schneiker S, Pühler A, Top EM. Schlüter A, et al. Plasmid. 2005 Sep;54(2):135-48. doi: 10.1016/j.plasmid.2005.03.001. Epub 2005 Apr 18. Plasmid. 2005. PMID: 16122561 - The Tcp conjugation system of Clostridium perfringens.
Wisniewski JA, Rood JI. Wisniewski JA, et al. Plasmid. 2017 May;91:28-36. doi: 10.1016/j.plasmid.2017.03.001. Epub 2017 Mar 7. Plasmid. 2017. PMID: 28286218 Review. - Spread and Persistence of Virulence and Antibiotic Resistance Genes: A Ride on the F Plasmid Conjugation Module.
Koraimann G. Koraimann G. EcoSal Plus. 2018 Jul;8(1):10.1128/ecosalplus.ESP-0003-2018. doi: 10.1128/ecosalplus.ESP-0003-2018. EcoSal Plus. 2018. PMID: 30022749 Free PMC article. Review.
Cited by
- The Transferable Resistome of Produce.
Blau K, Bettermann A, Jechalke S, Fornefeld E, Vanrobaeys Y, Stalder T, Top EM, Smalla K. Blau K, et al. mBio. 2018 Nov 6;9(6):e01300-18. doi: 10.1128/mBio.01300-18. mBio. 2018. PMID: 30401772 Free PMC article. - Evolution and dynamics of megaplasmids with genome sizes larger than 100 kb in the Bacillus cereus group.
Zheng J, Peng D, Ruan L, Sun M. Zheng J, et al. BMC Evol Biol. 2013 Dec 2;13:262. doi: 10.1186/1471-2148-13-262. BMC Evol Biol. 2013. PMID: 24295128 Free PMC article. - Alternative Ac/Ds transposition induces major chromosomal rearrangements in maize.
Zhang J, Yu C, Pulletikurti V, Lamb J, Danilova T, Weber DF, Birchler J, Peterson T. Zhang J, et al. Genes Dev. 2009 Mar 15;23(6):755-65. doi: 10.1101/gad.1776909. Genes Dev. 2009. PMID: 19299561 Free PMC article. - The IncP-6 plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions.
Haines AS, Jones K, Cheung M, Thomas CM. Haines AS, et al. J Bacteriol. 2005 Jul;187(14):4728-38. doi: 10.1128/JB.187.14.4728-4738.2005. J Bacteriol. 2005. PMID: 15995187 Free PMC article. - Complete Sequences of IncU Plasmids Harboring Quinolone Resistance Genes qnrS2 and aac(6')-Ib-cr in Aeromonas spp. from Ornamental Fish.
Dobiasova H, Videnska P, Dolejska M. Dobiasova H, et al. Antimicrob Agents Chemother. 2015 Nov 2;60(1):653-7. doi: 10.1128/AAC.01773-15. Print 2016 Jan. Antimicrob Agents Chemother. 2015. PMID: 26525788 Free PMC article.
References
- Allmeier, H., B. Cresnar, M. Greck, and R. Schmitt. 1992. Complete nucleotide-sequence of Tn1721—gene organization and a novel gene-product with features of a chemotaxis protein. Gene 111:11-20. - PubMed
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
- Austin, B., and C. A. Adams. 1996. Fish pathogens, p. 197-229. In B. Austin, M. Altwegg, P. J. Gosling, and S. Joseph (ed.), The genus Aeromonas. John Wiley & Sons, New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources