Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli - PubMed (original) (raw)
Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli
Iris Keren et al. J Bacteriol. 2004 Dec.
Abstract
Bacterial populations produce persisters, cells that neither grow nor die in the presence of bactericidal agents, and thus exhibit multidrug tolerance (MDT). The mechanisms of MDT and the nature of persisters have remained elusive. Our previous research has shown that persisters are largely responsible for the recalcitrance of biofilm infections. A general method for isolating persisters was developed, based on lysis of regular cells by ampicillin. A gene expression profile of persisters contained toxin-antitoxin (TA) modules and other genes that can block important cellular functions such as translation. Bactericidal antibiotics kill cells by corrupting the target function (for example, aminoglycosides interrupt translation, producing toxic peptides). We reasoned that inhibition of translation will lead to a shutdown of cellular functions, preventing antibiotics from corrupting their targets, giving rise to MDT persister cells. Overproduction of the RelE toxin, an inhibitor of translation, caused a sharp increase in persisters. Functional expression of a putative HipA toxin also increased persisters, while deletion of the hipBA module caused a sharp decrease in persisters in both stationary and biofilm populations. HipA is thus the first validated persister-MDT gene. We suggest that random fluctuation in the levels of MDT proteins leads to the formation of rare persister cells. The function of these specialized dormant cells is to ensure the survival of the population in the presence of lethal factors.
Figures
FIG. 1.
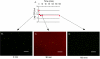
E. coli HM22 _hipA_7 cells were grown in LB medium to mid-exponential phase (∼5 × 107 cells/ml) at 37°C with aeration and treated with 50 μg of ampicillin/ml. (A) Samples were taken at indicated times and plated to determine live cells by colony count. Measurements were done in triplicate, and error bars indicate standard deviations. (B to D) Samples were stained with a LIVE/DEAD kit and visualized by epifluorescent microscopy (bar, 50 μm). Green cells are live, while red cells, stained by normally impermeant propidium iodide, are dead. Note the extensive red background due to cellular debris, as well as dead intact cells in the 30-min sample.
FIG. 2.
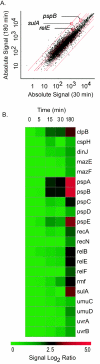
(A) Scatter plot of absolute gene expression at 180 min versus 30 min. Red lines indicate 2-, 3-, and 30-fold changes, respectively. (B) Heat map of selected genes from the cluster generated with Spotfire Decisionsite 7.2.
FIG. 3.
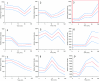
Cluster analysis of persister gene expression profile obtained with the Affymetrix Self-Organizing Map. The boxed cluster indicates the expression profile of genes specifically upregulated in persisters (180 min). The red lines indicate average signal intensity of all genes in the cluster, and blue lines indicate standard deviations.
FIG. 4.
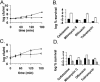
The effects of toxin overexpression on persister formation. (A) Cells were cultured as described in the legend to Fig. 1. RelE was induced (▪) from pKD3035 (pBAD::relE) in MG1 (MC1000 Δ_relBE_) at time zero by the addition of 0.2% arabinose, and MG1 with a blank vector (pBAD33) served as the control (▴). (B) Cells were cultured, and RelE was induced as described above. After 3 h of RelE induction, samples were removed and treated with either cefotaxime (100 μg/ml), mitomycin C (10 μg/ml), ofloxacin (5 μg ml), or tobramycin (25 μg/ml) for 3 h at 37°C with aeration. The control (MG1/pBAD) (black bars) was challenged at a cell density similar to that of the relE induced cells (white bars). (C) HM22 cells (K12 hipA7) were cultured as described above and at time zero were moved to 30°C (▪) or kept at 37°C (▴). (D) HM22 cells were challenged at 30°C (white bars) as described in panel B. Controls are HM22 (black bars) and HM21 (K-12 hipA wild type) (gray bars) cells were challenged at 37°C.
FIG. 5.
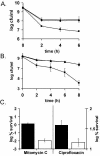
The effects of toxin deletion on persister formation. (A and B) EMG2 (K-12 wild type) (⧫), KL310 (Δ_relBE_) (▴) and KL312 (Δ_hipBA_) (▪) were grown to stationary phase (16 to 18 h) and challenged with ofloxacin (5 μg/ml) (A) or mitomycin C (10 μg/ml) (B). (C) Biofilms EMG2 (K-12 wild type) (black bars) and KL312 (Δ_hipBA_) (white bars) were grown at 37°C for 48 h on LB agar to ∼2 × 109 CFU/biofilm. They were then exposed to LB agar plates containing 20 mM NaNO3 with or without 5 μg of mitomycin C/ml (left y axis) and with or without 5 μg of ciprofloxacin/ml (right y axis). All experiments shown in Fig. 5 are an average of three independent measurements; error bars indicate standard deviations.
FIG. 6.
A model for tolerance and persister formation. (A) A toxin molecule such as an inhibitor of translation is produced, and cells that have a high level of this component due to random fluctuations of expression become persisters (B). (C) Antibiotic binds to its target (bottom), which generates a corrupted product (such as truncated peptides formed in the presence of aminoglycosides), leading to cell death. If a target is blocked by a toxic factor (top), then the antibiotic can bind, but will no longer corrupt the function, and the result is drug tolerance, allowing the cell to survive.
Similar articles
- Persister cells and the riddle of biofilm survival.
Lewis K. Lewis K. Biochemistry (Mosc). 2005 Feb;70(2):267-74. doi: 10.1007/s10541-005-0111-6. Biochemistry (Mosc). 2005. PMID: 15807669 Review. - Multidrug tolerance of biofilms and persister cells.
Lewis K. Lewis K. Curr Top Microbiol Immunol. 2008;322:107-31. doi: 10.1007/978-3-540-75418-3_6. Curr Top Microbiol Immunol. 2008. PMID: 18453274 Review. - GlpD and PlsB participate in persister cell formation in Escherichia coli.
Spoering AL, Vulic M, Lewis K. Spoering AL, et al. J Bacteriol. 2006 Jul;188(14):5136-44. doi: 10.1128/JB.00369-06. J Bacteriol. 2006. PMID: 16816185 Free PMC article. - Persister eradication: lessons from the world of natural products.
Keren I, Mulcahy LR, Lewis K. Keren I, et al. Methods Enzymol. 2012;517:387-406. doi: 10.1016/B978-0-12-404634-4.00019-X. Methods Enzymol. 2012. PMID: 23084949 - Persisters: a distinct physiological state of E. coli.
Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. Shah D, et al. BMC Microbiol. 2006 Jun 12;6:53. doi: 10.1186/1471-2180-6-53. BMC Microbiol. 2006. PMID: 16768798 Free PMC article.
Cited by
- Modeling single-cell phenotypes links yeast stress acclimation to transcriptional repression and pre-stress cellular states.
Bergen AC, Kocik RA, Hose J, McClean MN, Gasch AP. Bergen AC, et al. Elife. 2022 Nov 9;11:e82017. doi: 10.7554/eLife.82017. Elife. 2022. PMID: 36350693 Free PMC article. - Staphylococcus aureus in continuous culture: a tool for the rational design of antibiotic treatment protocols.
Udekwu KI, Levin BR. Udekwu KI, et al. PLoS One. 2012;7(7):e38866. doi: 10.1371/journal.pone.0038866. Epub 2012 Jul 20. PLoS One. 2012. PMID: 22911681 Free PMC article. - Association between toxin-antitoxin systems and biofilm formation.
Karimi S, Ghafourian S, Taheri Kalani M, Azizi Jalilian F, Hemati S, Sadeghifard N. Karimi S, et al. Jundishapur J Microbiol. 2014 Dec 8;8(1):e14540. doi: 10.5812/jjm.14540. eCollection 2015 Jan. Jundishapur J Microbiol. 2014. PMID: 25789127 Free PMC article. - Exploring antibiotic-induced persister formation and bacterial persistence genes in clinical isolates from Burkina Faso.
Konkobo A, Ouattara AK, Mètuor Dabiré A, Simporé J. Konkobo A, et al. BMC Infect Dis. 2024 Sep 17;24(1):994. doi: 10.1186/s12879-024-09906-9. BMC Infect Dis. 2024. PMID: 39289656 Free PMC article. - Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli.
Correia FF, D'Onofrio A, Rejtar T, Li L, Karger BL, Makarova K, Koonin EV, Lewis K. Correia FF, et al. J Bacteriol. 2006 Dec;188(24):8360-7. doi: 10.1128/JB.01237-06. Epub 2006 Oct 13. J Bacteriol. 2006. PMID: 17041039 Free PMC article.
References
- Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105-107. - PubMed
- Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. - PubMed
- Bayles, K. W. 2000. The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol. 8:274-278. - PubMed
- Bigger, J. W. 1944. Treatment of staphylococcal infections with penicillin. Lancet ii:497-500.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases