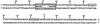Negative regulation of DNA repair gene (ung) expression by the CpxR/CpxA two-component system in Escherichia coli K-12 and induction of mutations by increased expression of CpxR - PubMed (original) (raw)
Negative regulation of DNA repair gene (ung) expression by the CpxR/CpxA two-component system in Escherichia coli K-12 and induction of mutations by increased expression of CpxR
Hiroshi Ogasawara et al. J Bacteriol. 2004 Dec.
Abstract
In Escherichia coli K-12 overexpressing CpxR, transcription of the ung gene for uracil-DNA glycosylase was repressed, ultimately leading to the induction of mutation. Gel shift, DNase I footprinting, and in vitro transcription assays all indicated negative regulation of ung transcription by phosphorylated CpxR. Based on the accumulated results, we conclude that ung gene expression is negatively regulated by the two-component system of CpxR/CpxA signal transduction.
Figures
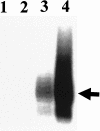
FIG. 1.
Overexpression of cpxR. The arrow indicates cpxR transcript from the lac promoter on the plasmid p41-5ΔGFP. BW25113(pCA24ΔGFP) (lanes 1 and 2) and BW25113(p41-5ΔGFP) (lanes 3 and 4) were grown to mid-log phase (OD600, 0.6) in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 1 mM IPTG. The S1 nuclease assay for the cpxR transcript was performed as described in Materials and Methods.
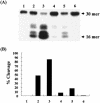
FIG. 2.
Assay of uracil-DNA glycosylase (Ung) activity. (A) Ung activity was assayed as described in Materials and Methods. For this assay, cell extracts (lanes 2 and 4, 25 ng; lanes 3 and 5, 50 ng) from BW25113(pCA24ΔGFP) (lanes 2 and 3), BW25113(p41-5ΔGFP) (lanes 4 and 5), and BWung (lane 6, 50 ng) were used. The 30-mer oligonucleotide was cleaved by Ung and NaOH to form a 16-mer product. In lane 1, no cell extract was used. (B) Cleaved DNA products (from panel A) were quantified with BAS 1000 Mac (Fuji film). Percent cleavage was determined by dividing the intensity of the cleaved product (16-mer) by the total intensity, which was defined as the sum of the intensities of the intact substrate and the cleaved products.
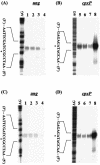
FIG. 3.
Effects of overexpressed CpxR or NlpE on ung and cpxP transcription. BW25113(pCA24ΔGFP) (A and B, lanes 1, 2, 5, and 6) and BW25113(p41-5ΔGFP) (A and B, lanes 3, 4, 7, and 8) were grown to mid-log phase (OD600, 0.6) in the absence (A and B, lanes 1, 3, 5, and 7) or presence (A and B, lanes 2, 4, 6, and 8) of 1 mM IPTG. S1 nuclease assays for ung (A) and cpxP (B) transcripts were performed as described in Materials and Methods. BW25113(pBAD18) (C and D, lanes 1, 2, 5, and 6) and BW25113(pBADnlpE) (C and D, lanes 3, 4, 7, and 8) were grown to mid-log phase (OD600, 0.6) in the absence (C and D, lanes 1, 3, 5, and 7) or presence (C and D, lanes 2, 4, 6, and 8) of 0.2% arabinose. After that, S1 nuclease assays for ung (C) and cpxP (D) transcripts were performed. Lane AG represents the Maxam-Gilbert sequence ladder. The transcription start site is marked with an asterisk.
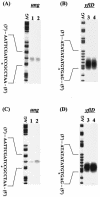
FIG. 4.
ung and yfiD gene expression dependent on growth phase. E. coli BW25113 (A and C, lane 1; B and D, lane 3) and BW27559 (Δ_cpxR_) (A and C, lane 2; B and D, lane 4) were grown to mid-log phase (OD600 = 0.6) (A, B) or early stationary phase (OD600 = 1.2) (C, D). After that, S1 nuclease assays for ung (A, C) and yfiD (B, D) transcripts were performed. Lane AG represents the Maxam-Gilbert sequence ladder. The transcription start site is marked with an asterisk.
FIG. 5.
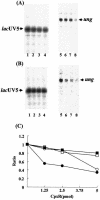
In vitro transcription assay. Single-round transcription in vitro of 0.1 pmol ung DNA template (−452 to +86 of promoter region) (A, lanes 5 to 8; B, lanes 5 to 8) or _lac_UV5 DNA template (A, lanes 1 to 4; B, lanes 1 to 4) was performed in the presence (B) or absence (A) of 10 mM acetylphosphate. The amounts of CpxR were as follows: lanes 1 and 5, 0 pmol; lanes 2 and 6, 1.25 pmol; lanes 3 and 7, 2.5 pmol; lanes 4 and 8, 5 pmol. Electrophoresis was performed with a 6% polyacrylamide sequencing gel. Bold arrows indicate the ung and _lac_UV5 transcripts. (C) ung (circles) and _lac_UV5 (square) transcripts in the absence (open symbols) and presence (closed symbols) of CpxR were quantified with BAS 1000 Mac (Fuji film). The relative value in shown as a ratio between each transcript and that in the absence of CpxR.
FIG. 6.
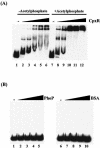
Gel shift assay. (A) Probe A was incubated at 37°C for 10 min with CpxR (lanes 1 to 6) or CpxR phosphorylated by acetylphosphate at 37°C (lanes 7 to 12). The amounts of CpxR were as follows: lanes 1 and 7, 0 pmol; lanes 2 and 8, 1.25 pmol; lanes 3 and 9, 2.5 pmol; lanes 4 and 10, 5 pmol; lanes 5 and 11, 7.5 pmol; lanes 6 and 12, 10 pmol. (B) Probe A was incubated at 37°C for 10 min with PhoP (lanes 1 to 5) or BSA (lanes 6 to 10) at 37°C. The amounts of PhoP or BSA were as follows: lanes 1 and 6, 0 pmol; lanes 2 and 7, 1.25 pmol; lanes 3 and 8, 2.5 pmol; lanes 4 and 9, 5 pmol; lanes 5 and 10, 10 pmol).
FIG. 7.
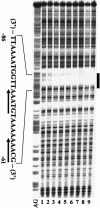
DNase I footprinting assay. Probe A was incubated with various amounts of the purified CpxR (lane 1, 0 pmol; lane 2, 10 pmol; lane 3, 20 pmol; lane 4, 30 pmol; lane 5, 40 pmol; lane 6, 50 pmol; lane 7, 60 pmol; lane 8, 70 pmol; lane 9, 80 pmol) and subjected to DNase I footprinting assays. Lanes AG represent the Maxam-Gilbert sequence ladder. The black boxes and bold arrows indicate the CpxR binding region and the direct repeat, respectively.
FIG. 8.
ung promoter and CpxR box. The transcription start site is marked with an asterisk. The nucleotide number represents the distance from the transcription initiation site of the ung promoter. Putative recognition sequences for σ70 (−10 and −35) are boxed. The arrows indicate the direct repeat in the CpxR box shown previously (6).
Similar articles
- The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins ompF and ompC.
Batchelor E, Walthers D, Kenney LJ, Goulian M. Batchelor E, et al. J Bacteriol. 2005 Aug;187(16):5723-31. doi: 10.1128/JB.187.16.5723-5731.2005. J Bacteriol. 2005. PMID: 16077119 Free PMC article. - Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli.
Yamamoto K, Ishihama A. Yamamoto K, et al. Biosci Biotechnol Biochem. 2006 Jul;70(7):1688-95. doi: 10.1271/bbb.60024. Biosci Biotechnol Biochem. 2006. PMID: 16861804 - The CpxR/CpxA two-component regulatory system up-regulates the multidrug resistance cascade to facilitate Escherichia coli resistance to a model antimicrobial peptide.
Weatherspoon-Griffin N, Yang D, Kong W, Hua Z, Shi Y. Weatherspoon-Griffin N, et al. J Biol Chem. 2014 Nov 21;289(47):32571-82. doi: 10.1074/jbc.M114.565762. Epub 2014 Oct 6. J Biol Chem. 2014. PMID: 25294881 Free PMC article. - [Cpx two-component regulatory system in gram-negative bacteria--a review].
Xu L, Zhou X, He X. Xu L, et al. Wei Sheng Wu Xue Bao. 2014 Mar 4;54(3):269-75. Wei Sheng Wu Xue Bao. 2014. PMID: 24984518 Review. Chinese.
Cited by
- The Two-Component System CpxRA Represses Salmonella Pathogenicity Island 2 by Directly Acting on the ssrAB Regulatory Operon.
León-Montes N, Nava-Galeana J, Rodríguez-Valverde D, Soria-Bustos J, Rosales-Reyes R, Rivera-Gutiérrez S, Hirakawa H, Ares MA, Bustamante VH, De la Cruz MA. León-Montes N, et al. Microbiol Spectr. 2022 Oct 26;10(5):e0271022. doi: 10.1128/spectrum.02710-22. Epub 2022 Sep 8. Microbiol Spectr. 2022. PMID: 36073960 Free PMC article. - Cyclic di-GMP Positively Regulates DNA Repair in Vibrio cholerae.
Fernandez NL, Srivastava D, Ngouajio AL, Waters CM. Fernandez NL, et al. J Bacteriol. 2018 Jul 10;200(15):e00005-18. doi: 10.1128/JB.00005-18. Print 2018 Aug 1. J Bacteriol. 2018. PMID: 29610212 Free PMC article. - A complex transcription network controls the early stages of biofilm development by Escherichia coli.
Prüss BM, Besemann C, Denton A, Wolfe AJ. Prüss BM, et al. J Bacteriol. 2006 Jun;188(11):3731-9. doi: 10.1128/JB.01780-05. J Bacteriol. 2006. PMID: 16707665 Free PMC article. Review. No abstract available. - A genome-wide MeSH-based literature mining system predicts implicit gene-to-gene relationships and networks.
Xiang Z, Qin T, Qin ZS, He Y. Xiang Z, et al. BMC Syst Biol. 2013 Oct 16;7 Suppl 3(Suppl 3):S9. doi: 10.1186/1752-0509-7-S3-S9. BMC Syst Biol. 2013. PMID: 24555475 Free PMC article. - Characterization of the Cpx regulon in Escherichia coli strain MC4100.
Price NL, Raivio TL. Price NL, et al. J Bacteriol. 2009 Mar;191(6):1798-815. doi: 10.1128/JB.00798-08. Epub 2008 Dec 19. J Bacteriol. 2009. PMID: 19103922 Free PMC article.
References
- Danese, P. N., and T. J. Silhavy. 1997. The σE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 11:1183-1193. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases