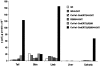Transgenic mice expressing a ligand-inducible cre recombinase in osteoblasts and odontoblasts: a new tool to examine physiology and disease of postnatal bone and tooth - PubMed (original) (raw)
Transgenic mice expressing a ligand-inducible cre recombinase in osteoblasts and odontoblasts: a new tool to examine physiology and disease of postnatal bone and tooth
Jung-Eun Kim et al. Am J Pathol. 2004 Dec.
Abstract
The skeleton supports body structures in vertebrates and helps maintain calcium homeostasis throughout life. Disruption of genes involved in mammalian bone formation has often led to embryonic lethality, hence preventing study of these genes' role in adult animals. To develop a usable tool for such study, we generated transgenic mice in which a 2.3-kb mouse Col1a1 proximal promoter, which is active in all osteoblasts, drives a transgene coding for a polypeptide consisting of Cre recombinase fused to a mutated ligand-binding domain of the estrogen receptor. In this Col1a1-CreERT2 mouse line, expression patterns of the transgene and of the resulting Cre-mediated DNA recombination are analyzed by crossing with ROSA26 reporter mice and by measurement of beta-galactosidase activity and X-gal staining. Exposure to 4-hydroxytamoxifen induced Cre-mediated recombination in osteoblasts in virtually all bones and in odontoblasts in teeth of both embryos and postnatal mice. The generation of these transgenic mice provides a new and important tool with which to study the function of specific genes in bone and tooth physiology and diseases in intact animals after birth.
Figures
Figure 1
Generation of the Col1a1-CreERT2 transgene. A: Construction of the Col1a1-CreERT2 transgene for osteoblast-specific expression of ligand-dependent Cre recombinase. The Col1a1 promoter cassette contains a 2.3-kb proximal fragment of the mouse type I collagen promoter. The ERT2 LBD of the ER contains the G400V/M543A/L544A triple mutation. B: RT-PCR analysis of Cre mRNA expression in limbs of embryos at 13.5, 16.5, and 18.5 dpc and of 3-day-old pups. Mouse GAPDH PCR shows that the same amount of cDNA was used in each sample. Wt and TG indicate wild-type and Col1a1-CreERT2 transgenic embryo or pup, respectively.
Figure 2
Inducible Cre-mediated β-gal activity in 18-day-old Col1a1-CreERT2;R26R double-transgenic mice. Pups harboring both the Col1a1-CreERT2 transgene and the ROSA26-LacZ locus had been injected intraperitoneally with 0.75 mg of 4-OHT for 5 consecutive days starting at 12 days of age and then killed 48 hours later. The indicated tissues were homogenized and assayed for β-gal activity using a chemiluminescence assay as described in Materials and Methods. Wt indicates wild-type mice. Enzymatic activity is presented as mean relative light units (RLU) divided by the protein concentration.
Figure 3
LacZ expression induced by 4-OHT in whole-mount transgenic embryos and postnatal mice. A: Eighteen-day-old pups containing the Col1a1-CreERT2;R26R double transgene had been injected intraperitoneally with 1 mg of 4-OHT for 5 consecutive days starting at 12 days of age and were killed 48 hours later. B: Control pups were not injected with 4-OHT. X-Gal staining was strong in the bones of the skull (A1), calvaria (A2), digits (A3), and axial skeleton (A4) of 4-OHT-injected pups but weak in the corresponding bones of control pups (B1–B4). C: Female R26R mice were mated with male Col1a1-CreERT2 mice, injected intraperitoneally three times with 1 mg of 4-OHT or sunflower oil starting at 14.5 dpc, and then killed 48 hours later; the 18.5 dpc embryos were stained with X-gal. Only the Col1a1-CreERT2;R26R double-transgenic embryos from 4-OHT-treated dams show LacZ expression in bone (arrowheads). Higher-magnification images of the cranial view of the skull (D1 and E1), caudal view of the skull (D2 and E2), forelimbs (D3 and E3), vertebrae (D4 and E4), and ribs (D5 and E5) of 18.5 dpc double-transgenic embryos whose dams were treated with 4-OHT or sunflower oil, respectively.
Figure 4
Histological analysis of bones of Col1a1-CreERT2;R26R double-transgenic embryos and pups. Analysis was performed after whole-mount staining with X-gal and counterstaining with nuclear fast red. A and B: Histological results for 18.5 dpc Col1a1-CreERT2;R26R embryos whose dams had been treated with 4-OHT or sunflower oil embryos, respectively. A1, A2, B1, and B2: Coronal sections of the skull. Arrowheads indicate strong X-gal staining in osteoblasts of calvaria and jaw. A3 and B3: Longitudinal sections of the femur. A4 and B4: Higher-magnification images corresponding to the boxed areas in A3 and B3, respectively. In embryos whose dams had been treated with 4-OHT, no X-gal staining was detected in the heart (C1), lung (C2), liver (C3), or kidney (C4). D and E: Histological results of longitudinal section of femur from 18-day-old Col1a1-CreERT2;R26R pups with or without treatment with 4-OHT, respectively. X-Gal staining shows osteoblasts of femur. D2, D4, E2, and E4: Higher-magnification images corresponding to of the boxed areas in D1, D3, E1, and E3, respectively. B and BM in D3 and E3 indicate bone and bone marrow cavity, respectively. In pups treated with 4-OHT, no X-gal staining was detected in the skin (F1), liver (F2), or kidney (F3).
Figure 5
Histological analysis of coronal sections of molars and incisors of Col1a1-CreERT2;R26R double-transgenic 18.5 dpc embryos (A and B) and 18-day-old mice (C and D). Staining was performed with X-gal and counterstaining with nuclear fast red. X-Gal staining was strong in the odontoblasts of molar and incisor. A and C: Histological results for the Col1a1-CreERT2;R26R embryos and pups treated with 4-OHT, respectively. B and D: Histological results for the Col1a1-CreERT2;R26R embryos and pups not treated with 4-OHT, respectively. Columns 2 and 4 are higher magnification images corresponding to columns 1 and 3, respectively.
Similar articles
- Odontoblast-specific expression of cre recombinase successfully deletes gene segments flanked by loxP sites in mouse teeth.
Sreenath TL, Cho A, Thyagarajan T, Kulkarni AB. Sreenath TL, et al. Genesis. 2003 Feb;35(2):94-9. doi: 10.1002/gene.10170. Genesis. 2003. PMID: 12533791 - Mouse alpha1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast.
Dacquin R, Starbuck M, Schinke T, Karsenty G. Dacquin R, et al. Dev Dyn. 2002 Jun;224(2):245-51. doi: 10.1002/dvdy.10100. Dev Dyn. 2002. PMID: 12112477 - Controlling gene expression in the urothelium using transgenic mice with inducible bladder specific Cre-lox recombination.
Bex A, Vooijs M, Horenblas S, Berns A. Bex A, et al. J Urol. 2002 Dec;168(6):2641-4. doi: 10.1016/S0022-5347(05)64235-8. J Urol. 2002. PMID: 12442001 - Endothelial-Specific Cre Mouse Models.
Payne S, De Val S, Neal A. Payne S, et al. Arterioscler Thromb Vasc Biol. 2018 Nov;38(11):2550-2561. doi: 10.1161/ATVBAHA.118.309669. Arterioscler Thromb Vasc Biol. 2018. PMID: 30354251 Free PMC article. Review. - Sites of Cre-recombinase activity in mouse lines targeting skeletal cells.
Couasnay G, Madel MB, Lim J, Lee B, Elefteriou F. Couasnay G, et al. J Bone Miner Res. 2021 Sep;36(9):1661-1679. doi: 10.1002/jbmr.4415. Epub 2021 Aug 22. J Bone Miner Res. 2021. PMID: 34278610 Review.
Cited by
- Optimizing tamoxifen-inducible Cre/loxp system to reduce tamoxifen effect on bone turnover in long bones of young mice.
Zhong ZA, Sun W, Chen H, Zhang H, Lay YE, Lane NE, Yao W. Zhong ZA, et al. Bone. 2015 Dec;81:614-619. doi: 10.1016/j.bone.2015.07.034. Epub 2015 Jul 29. Bone. 2015. PMID: 26232373 Free PMC article. - Sample preparation for high-resolution 3D confocal imaging of mouse skeletal tissue.
Kusumbe AP, Ramasamy SK, Starsichova A, Adams RH. Kusumbe AP, et al. Nat Protoc. 2015 Dec;10(12):1904-14. doi: 10.1038/nprot.2015.125. Epub 2015 Oct 29. Nat Protoc. 2015. PMID: 26513669 - Comparative Evaluation of Inducible Cre Mouse Models for Fibroblast Targeting in the Healthy and Infarcted Myocardium.
Aguado-Alvaro LP, Garitano N, Abizanda G, Larequi E, Prosper F, Pelacho B. Aguado-Alvaro LP, et al. Biomedicines. 2022 Sep 21;10(10):2350. doi: 10.3390/biomedicines10102350. Biomedicines. 2022. PMID: 36289614 Free PMC article. - Kruppel-like Factors in Skeletal Physiology and Pathologies.
Abe M, Saeki N, Ikeda Y, Ohba S. Abe M, et al. Int J Mol Sci. 2022 Dec 2;23(23):15174. doi: 10.3390/ijms232315174. Int J Mol Sci. 2022. PMID: 36499521 Free PMC article. Review. - Atf7ip Inhibits Osteoblast Differentiation via Negative Regulation of the Sp7 Transcription Factor.
Hu G, Shi X, Qu X, Han C, Hu A, Jia Z, Yang J, Liu H, Wu Y. Hu G, et al. Int J Mol Sci. 2023 Feb 21;24(5):4305. doi: 10.3390/ijms24054305. Int J Mol Sci. 2023. PMID: 36901736 Free PMC article.
References
- Aszodi A, Bateman JF, Gustafsson E, Boot-Handford R, Fassler R. Mammalian skeletogenesis and extracellular matrix: what can we learn from knockout mice? Cell Struct Funct. 2000;25:71–82. - PubMed
- Mundlos S, Olsen BR. Heritable diseases of the skeleton. Part I: molecular insights into skeletal development-transcription factors and signaling pathways. FASEB J. 1997;11:125–132. - PubMed
- Mundlos S, Olsen BR. Heritable diseases of the skeleton. Part II: molecular insights into skeletal development-matrix components and their homeostasis. FASEB J. 1997;11:227–233. - PubMed
- Capecchi MR. The new mouse genetics: altering the genome by gene targeting. Trends Genet. 1989;5:70–76. - PubMed
- Rossant J, McMahon A. “Cre”-ating mouse mutants—a meeting review on conditional mouse genetics. Genes Dev. 1999;13:142–145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous