Probing conformational changes of human serum albumin due to unsaturated fatty acid binding by chemical cross-linking and mass spectrometry - PubMed (original) (raw)
Probing conformational changes of human serum albumin due to unsaturated fatty acid binding by chemical cross-linking and mass spectrometry
Bill X Huang et al. Biochem J. 2005.
Abstract
Mass spectrometry with chemical cross-linking was used to probe the conformational changes of HSA (human serum albumin) in solution on interaction with monounsaturated OA (oleic acid) or polyunsaturated AA (arachidonic acid) or DHA (docosahexaenoic acid). Fatty acid-free or -bound HSA was modified with lysine-specific cross-linkers and digested with trypsin. Cross-linked peptides were analysed by nano-electrospray ionization MS to localize the sites of cross-linking. Our data indicated that a local conformational change involving movement of the side chains of Lys-402 of subdomain IIIA or Lys-541 of subdomain IIIB occurred upon binding of all three fatty acids. Our data also indicated that the side chains of Lys-205 (IIA) and Lys-466 (IIIA) moved closer towards each other upon binding AA or DHA, but not OA, suggesting that the conformations of HSA when bound to mono- and poly-unsaturated fatty acids are distinctively different. While these observations agreed with previous X-ray crystallographic studies, the distances between epsilon-amino groups of most cross-linked lysine pairs were shorter than the crystal structure predicted, possibly reflecting a discrepancy between the solution and crystal structures. This method can serve as a useful complement to X-ray crystallography, particularly in probing the structure of a protein in solution.
Figures
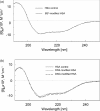
Figure 1. Far-UV CD spectra of BS3-modified (a) and DSS- or DSG-modified (b) HSA
DSS- and DSG-modified samples were dialysed against pure water using a membrane with molecular mass cut-off of 3500 Da before CD measurements.
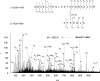
Figure 2. Nano-ESI-QqTOF reconstructed mass spectra of tryptic digests from unmodified (a), and BS3-modified control (b), OA-bound (c), AA-bound (d) and DHA-bound (e) HSA
The peak with a mass of 3552.8 Da emerged from BS3-modified samples (b–e). The peak with a mass of 4505 Da emerged from AA- or DHA-bound HSA samples (d, e), but was absent from OA-bound HSA (c). Cross-linked peptides in the fatty acid-free control (b) are marked with an asterisk (*).
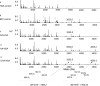
Figure 3. Nano-ESI-QqTOF-MS/MS analysis of the cross-linked peptide with mass of 3552.8 Da depicted in Figure 2
The sequence of the peptide is shown using the single-letter code, based on the fragmentation observed in the spectrum. The fragments in the spectra indicated two peptide segments, Q[390–410]R (Gln-390–Arg-410) and A[539–545]K (Ala-539–Lys-545), cross-linked at Lys-402/Lys-541. The peptide segments are labelled α and β respectively according to the nomenclature suggested by Schilling et al. [30]. N-terminal b ions and C-terminal y ions resulting from the amide bond cleavages are labelled.
Figure 4. Nano-ESI-QqTOF reconstructed mass spectra of BS3-modified HSA samples
(a) Fatty acid-free sample digested in H218O; (b) mixture of the DHA-bound sample digested in normal H216O and the fatty acid-free sample digested in H18O (at a ratio of 1:2).
Figure 5. Extracted ion chromatograms of a cross-linked peptide obtained from DSG-modified (left) and DSS-modified (right) HSA samples analysed by HPLC/nano-ESI/MS/MS
(a) Fatty acid-free control; (b) OA-bound HSA; (c) AA-bound HSA; (d) DHA-bound HSA. A new peak with a mass of 4463 Da (left) or 4505 Da (right), derived from the quintuply charged ion at m/z of 893.6 or 902.0 respectively, eluted at around 43.5 min with AA- or DHA-bound HSA (c, d).
Figure 6. Conformational changes of HSA induced by fatty acid binding depicted based on the crystal structure of HSA complexed with AA
The rotation of the side chains of Lys-402 (domain IIIA) and Lys-541 (domain IIIB) towards each other upon binding to a fatty acid is suggested. In addition, depicted is the movement of the side chains of Lys-205 from domain IIA and Lys-466 from domain IIIA closer towards each other upon binding of polyunsaturated AA or DHA. The cross-linking resulting from these movements is indicated. Fatty acid molecules are represented in space-filling form, and their binding sites shown in the right panel are based on the PDB entry 1gnj.
Similar articles
- Crystal structures of human serum albumin complexed with monounsaturated and polyunsaturated fatty acids.
Petitpas I, Grüne T, Bhattacharya AA, Curry S. Petitpas I, et al. J Mol Biol. 2001 Dec 14;314(5):955-60. doi: 10.1006/jmbi.2000.5208. J Mol Biol. 2001. PMID: 11743713 - Probing three-dimensional structure of bovine serum albumin by chemical cross-linking and mass spectrometry.
Huang BX, Kim HY, Dass C. Huang BX, et al. J Am Soc Mass Spectrom. 2004 Aug;15(8):1237-47. doi: 10.1016/j.jasms.2004.05.004. J Am Soc Mass Spectrom. 2004. PMID: 15276171 - A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions.
Buttar D, Colclough N, Gerhardt S, MacFaul PA, Phillips SD, Plowright A, Whittamore P, Tam K, Maskos K, Steinbacher S, Steuber H. Buttar D, et al. Bioorg Med Chem. 2010 Nov 1;18(21):7486-96. doi: 10.1016/j.bmc.2010.08.052. Epub 2010 Sep 24. Bioorg Med Chem. 2010. PMID: 20869876 - Determination of hexahydrophthalic anhydride adducts to human serum albumin.
Kristiansson MH, Lindh CH, Jönsson BA. Kristiansson MH, et al. Biomarkers. 2003 Sep-Oct;8(5):343-59. doi: 10.1080/13547500310001607836. Biomarkers. 2003. PMID: 14602520 - Molecular dynamics study of conformational changes in human serum albumin by binding of fatty acids.
Fujiwara S, Amisaki T. Fujiwara S, et al. Proteins. 2006 Aug 15;64(3):730-9. doi: 10.1002/prot.21053. Proteins. 2006. PMID: 16783783
Cited by
- The role of the protein-binding on the mode of drug action as well the interactions with other drugs.
Tesseromatis C, Alevizou A. Tesseromatis C, et al. Eur J Drug Metab Pharmacokinet. 2008 Oct-Dec;33(4):225-30. doi: 10.1007/BF03190876. Eur J Drug Metab Pharmacokinet. 2008. PMID: 19230595 Review. - The Antistaphylococcal Lysin, CF-301, Activates Key Host Factors in Human Blood To Potentiate Methicillin-Resistant Staphylococcus aureus Bacteriolysis.
Indiani C, Sauve K, Raz A, Abdelhady W, Xiong YQ, Cassino C, Bayer AS, Schuch R. Indiani C, et al. Antimicrob Agents Chemother. 2019 Mar 27;63(4):e02291-18. doi: 10.1128/AAC.02291-18. Print 2019 Apr. Antimicrob Agents Chemother. 2019. PMID: 30670427 Free PMC article. - Biochemical and biological functions of docosahexaenoic acid in the nervous system: modulation by ethanol.
Kim HY. Kim HY. Chem Phys Lipids. 2008 May;153(1):34-46. doi: 10.1016/j.chemphyslip.2008.02.014. Epub 2008 Mar 2. Chem Phys Lipids. 2008. PMID: 18359292 Free PMC article. Review. - Antigenic changes in human albumin caused by reactivity with the occupational allergen diphenylmethane diisocyanate.
Wisnewski AV, Liu J, Redlich CA. Wisnewski AV, et al. Anal Biochem. 2010 May 15;400(2):251-8. doi: 10.1016/j.ab.2010.01.037. Epub 2010 Feb 1. Anal Biochem. 2010. PMID: 20123080 Free PMC article. - High-Density Lipoprotein Biogenesis: Defining the Domains Involved in Human Apolipoprotein A-I Lipidation.
Pollard RD, Fulp B, Sorci-Thomas MG, Thomas MJ. Pollard RD, et al. Biochemistry. 2016 Sep 6;55(35):4971-81. doi: 10.1021/acs.biochem.6b00347. Epub 2016 Aug 23. Biochemistry. 2016. PMID: 27501467 Free PMC article.
References
- Carter D. C., He X., Munson S. H., Twigg P. D., Gernert K. M., Broom M. B., Miller T. Y. Three-dimensional structure of human serum albumin. Science. 1989;244:1195–1198. - PubMed
- Carter D. C., Ho J. X. Structure of serum albumin. Adv. Protein Chem. 1994;45:153–203. - PubMed
- Spector A. Fatty acid binding to plasma albumin. J. Lipid Res. 1975;16:165–179. - PubMed
- He X. M., Carter D. C. Atomic structure and chemistry of human serum albumin. Nature (London) 1992;358:209–215. - PubMed
- Sugio S., Kashima A., Mochizuki S., Noda M., Kobayashi K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999;12:439–446. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources