Status of the microbial census - PubMed (original) (raw)
Review
Status of the microbial census
Patrick D Schloss et al. Microbiol Mol Biol Rev. 2004 Dec.
Abstract
Over the past 20 years, more than 78,000 16S rRNA gene sequences have been deposited in GenBank and the Ribosomal Database Project, making the 16S rRNA gene the most widely studied gene for reconstructing bacterial phylogeny. While there is a general appreciation that these sequences are largely unique and derived from diverse species of bacteria, there has not been a quantitative attempt to describe the extent of sequencing efforts to date. We constructed rarefaction curves for each bacterial phylum and for the entire bacterial domain to assess the current state of sampling and the relative taxonomic richness of each phylum. This analysis quantifies the general sense among microbiologists that we are a long way from a complete census of the bacteria on Earth. Moreover, the analysis indicates that current sampling strategies might not be the most effective ones to describe novel diversity because there remain numerous phyla that are globally distributed yet poorly sampled. Based on the current level of sampling, it is not possible to estimate the total number of bacterial species on Earth, but the minimum species richness is 35,498. Considering previous global species richness estimates of 10(7) to 10(9), we are certain that this estimate will increase with additional sequencing efforts. The data support previous calls for extensive surveys of multiple chemically disparate environments and of specific phylogenetic groups to advance the census most rapidly.
Figures
FIG. 1.
Phylogenetic tree of the Bacteria showing established phyla (italicized Latinized names) and candidate phyla described previously (9, 10, 19), using the November 2003 ARB database (
[15]) with 16,964 sequences that are over 1,000 bp. The vertex angle of each wedge indicates the relative abundance of sequences in each phylum, and the length of each side of the wedge indicates the range of branching depth found in that phylum. The density of shading of each wedge corresponds to the proportion of sequences in that phylum obtained from cultured representatives. None of the candidate phyla have cultured representatives.
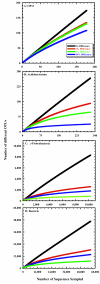
FIG.2.
Rarefaction curves constructed with accessions from the RDP-II in the OP11 (A), Acidobacterium (B), and γ-Proteobacteria (C) phyla and for all 16S rRNA genes (D) at various distances. Each distance represents the maximum difference allowed for DOTUR to consider a group of sequences to be in the same OTU.
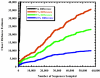
FIG. 3.
Collector's curve of the Chao1 nonparametric richness estimator for sequences in the RDP-II. Accession numbers were used to determine the order in which sequences have been sampled. OTUs defined by a collection of identical sequences reached an estimate of 325,040 different OTUs.
References
- Baker, B. J., G. W. Tyson, P. Hugenholtz, and J. F. Banfield. 2004. Analysis of genomic shotgun sequence data from an acid mine drainage biofilm community reveals a novel Euryarchaeota. Abstr. 104th Gen. Meet. Am. Soc. Microbiol. 2004, poster 161.
- Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270.
- Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous