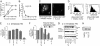Potent inhibition of huntingtin aggregation and cytotoxicity by a disulfide bond-free single-domain intracellular antibody - PubMed (original) (raw)
Potent inhibition of huntingtin aggregation and cytotoxicity by a disulfide bond-free single-domain intracellular antibody
David W Colby et al. Proc Natl Acad Sci U S A. 2004.
Erratum in
- Proc Natl Acad Sci U S A. 2005 Jan 18;102(3):955
Abstract
Huntington's disease (HD) is a progressive neurodegenerative disorder caused by an expansion in the number of polyglutamine-encoding CAG repeats in the gene that encodes the huntingtin (htt) protein. A property of the mutant protein that is intimately involved in the development of the disease is the propensity of the glutamine-expanded protein to misfold and generate an N-terminal proteolytic htt fragment that is toxic and prone to aggregation. Intracellular antibodies (intrabodies) against htt have been shown to reduce htt aggregation by binding to the toxic fragment and inactivating it or preventing its misfolding. Intrabodies may therefore be a useful gene-therapy approach to treatment of the disease. However, high levels of intrabody expression have been required to obtain even limited reductions in aggregation. We have engineered a single-domain intracellular antibody against htt for robust aggregation inhibition at low expression levels by increasing its affinity in the absence of a disulfide bond. Furthermore, the engineered intrabody variable light-chain (V(L))12.3, rescued toxicity in a neuronal model of HD. We also found that V(L)12.3 inhibited aggregation and toxicity in a Saccharomyces cerevisiae model of HD. V(L)12.3 is significantly more potent than earlier anti-htt intrabodies and is a potential candidate for gene therapy treatment for HD. To our knowledge, this is the first attempt to improve affinity in the absence of a disulfide bond to improve intrabody function. The demonstrated importance of disulfide bond-independent binding for intrabody potency suggests a generally applicable approach to the development of effective intrabodies against other intracellular targets.
Figures
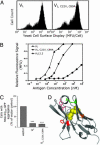
Fig. 1.
A single-domain intrabody against htt was engineered for high affinity in the absence of a disulfide bond. (A) Histograms of yeast cell-surface expression levels for VL and VL,C22V,C89A, indicating comparable levels of expression with and without the disulfide bond. mfu, mean fluorescence units. (B) Antigen-binding curves for YSD VL mutants measured by flow cytometry. Values normalized to maximal intensity were measured, except for VL,C22V,C89A, which was normalized to maximal intensity measured for VL.VL (♦) has a _K_d of ≈30 nM, whereas VL with cysteine mutations (VL,C22V,C89A; •) has significantly lower binding affinity (>10 μM). Repeated rounds of random mutagenesis of VL,C22V,C89A, followed by sorting for improved binding resulted in the mutant VL12.3, which has a _K_d of ≈3 nM. (C) Effect of VL and VL,C22V,C89A on htt aggregation in ST14A cells transiently transfected with indicated intrabody or vector control and httex1Q97-GFP at a 2:1 plasmid ratio. Both intrabodies are equally capable of partially blocking aggregation when overexpressed at high levels. ***, P < 0.001. (D) Homology model showing mutations obtained during engineering; the model contains residues present before mutagenesis. Cysteine residues are yellow. Mutations observed after mutagenesis and sorting are F37I (red), Y51D (pink), K67R (green), and A75T (orange).
Fig. 2.
Engineered VL12.3 robustly blocks htt aggregation in several different cell lines. (A) ST14A cells were transiently cotransfected with httex1Q97-GFP and either an intrabody [C4 (3) or VL12.3] or an empty control vector, and cells with visible aggregates were counted 1, 2, and 3 days posttransfection (5:1 intrabody:htt plasmid ratio, n = 3). VL12.3 (•) persistently eliminated htt aggregation over 3 days. (B) Dose–response of VL12.3 was measured at 2 days by using various intrabody:htt plasmid ratios (n = 3). (C) Fluorescence microscopy images of typical cells. (D) Flow cytometry histograms showing expression level per cell of httex1Q97-GFP in transfected cells in the presence of intrabody compared with empty vector (mean fluorescence intensity 82 vs. 76 mfu, respectively; transfection efficiencies were comparable in both samples, at 13% and 11%, respectively). (E) Comparison of intrabody activity for the intrabodies mentioned above and a non-htt binding intrabody (scFv ML3.9) and wild-type VL at a 1:1 intrabody:htt plasmid ratio (***, P < 0.001) in SH-SY5Y human neuroblastoma cells. (F) Partial dose–response for the same intrabodies in HEK293 cells. (G) Western blot of Triton X-100-soluble and -insoluble fractions of cells lysed 24 h after cotransfection at a 2:1 intrabody:htt ratio. (H) Anti-His6 Western blot of intrabody expression levels in transiently transfected HEK293 cells.
Fig. 3.
Engineered intrabody VL12.3 inhibits metabolic dysfunction in neuronal model of HD. ST14A cells were transfected with a plasmid encoding GFP, httex1Q25-GFP, httex1Q97-GFP, or httex1Q97-GFP with VL12.3 in a 2:1 ratio. (A) Live GFP-positive cells were collected by FACS in a 96-well plate, 35,000 cells per well, at 48 h posttransfection; typical dot plot is shown for a GFP sample. Other samples showed a similar pattern, and the sorting gate (boxed area) was the same in all instances. (B). Cells were incubated with MTT reagent for 3 h, solubilized, and the A570 was measured; mean values from three separate experiments containing all four samples are shown. Statistics directly over error bars are for comparison with GFP. ns, not significant; *, P < 0.05; **, P < 0.01. Statistics over brackets are comparisons between the two samples indicated. Four additional pairwise comparisons may be made between httex1Q97-GFP and httex1Q97-GFP plus VL12.3; the pooled results indicate a 56 ± 25% increase in A570; P < 0.001. Expression of httex1Q97-GFP significantly reduced the ability of cells to reduce MTT, but this effect was reversed by the coexpression of VL12.3.
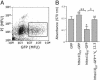
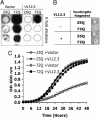
Fig. 4.
VL12.3 suppresses aggregation and rescues toxicity in an S. cerevisiae model of HD. (A) Filter retardation assay showing httex1Q72-CFP aggregates (dark) from lysates of cells expressing httex1Q25-CFP or httex1Q72-CFP with either VL12.3 or an empty vector control. Dashed circles indicate where insoluble material would appear. Difference between 25Q with and without VL12.3 is insignificant and within the variance usually observed for the assay. (B) Spottings of yeast strains, indicating ability to grow on solid media. (C) Growth curves obtained by measuring the OD600 of yeast cultures. Yeast expressing VL12.3-YFP along with httex1Q72-CFP grow at rates comparable with those expressing htt with nonpathological polyglutamine repeat lengths, in contrast to those carrying an empty vector only.
Similar articles
- Effects of intracellular expression of anti-huntingtin antibodies of various specificities on mutant huntingtin aggregation and toxicity.
Khoshnan A, Ko J, Patterson PH. Khoshnan A, et al. Proc Natl Acad Sci U S A. 2002 Jan 22;99(2):1002-7. doi: 10.1073/pnas.022631799. Epub 2002 Jan 15. Proc Natl Acad Sci U S A. 2002. PMID: 11792860 Free PMC article. - Development of a human light chain variable domain (V(L)) intracellular antibody specific for the amino terminus of huntingtin via yeast surface display.
Colby DW, Garg P, Holden T, Chao G, Webster JM, Messer A, Ingram VM, Wittrup KD. Colby DW, et al. J Mol Biol. 2004 Sep 17;342(3):901-12. doi: 10.1016/j.jmb.2004.07.054. J Mol Biol. 2004. PMID: 15342245 - Green tea (-)-epigallocatechin-gallate modulates early events in huntingtin misfolding and reduces toxicity in Huntington's disease models.
Ehrnhoefer DE, Duennwald M, Markovic P, Wacker JL, Engemann S, Roark M, Legleiter J, Marsh JL, Thompson LM, Lindquist S, Muchowski PJ, Wanker EE. Ehrnhoefer DE, et al. Hum Mol Genet. 2006 Sep 15;15(18):2743-51. doi: 10.1093/hmg/ddl210. Epub 2006 Aug 7. Hum Mol Genet. 2006. PMID: 16893904 - Engineered antibody therapies to counteract mutant huntingtin and related toxic intracellular proteins.
Butler DC, McLear JA, Messer A. Butler DC, et al. Prog Neurobiol. 2012 May;97(2):190-204. doi: 10.1016/j.pneurobio.2011.11.004. Epub 2011 Nov 18. Prog Neurobiol. 2012. PMID: 22120646 Free PMC article. Review. - [Huntington disease. A review].
Bonilla E. Bonilla E. Invest Clin. 2000 Jun;41(2):117-41. Invest Clin. 2000. PMID: 10961047 Review. Spanish.
Cited by
- Huntingtin aggregation kinetics and their pathological role in a Drosophila Huntington's disease model.
Weiss KR, Kimura Y, Lee WC, Littleton JT. Weiss KR, et al. Genetics. 2012 Feb;190(2):581-600. doi: 10.1534/genetics.111.133710. Epub 2011 Nov 17. Genetics. 2012. PMID: 22095086 Free PMC article. - Evidence for prion-like mechanisms in several neurodegenerative diseases: potential implications for immunotherapy.
Marciniuk K, Taschuk R, Napper S. Marciniuk K, et al. Clin Dev Immunol. 2013;2013:473706. doi: 10.1155/2013/473706. Epub 2013 Oct 20. Clin Dev Immunol. 2013. PMID: 24228054 Free PMC article. Review. - An scFv intrabody against the nonamyloid component of alpha-synuclein reduces intracellular aggregation and toxicity.
Lynch SM, Zhou C, Messer A. Lynch SM, et al. J Mol Biol. 2008 Mar 14;377(1):136-47. doi: 10.1016/j.jmb.2007.11.096. Epub 2007 Dec 5. J Mol Biol. 2008. PMID: 18237741 Free PMC article. - Targeted Intracellular Delivery of Antibodies: The State of the Art.
Slastnikova TA, Ulasov AV, Rosenkranz AA, Sobolev AS. Slastnikova TA, et al. Front Pharmacol. 2018 Oct 24;9:1208. doi: 10.3389/fphar.2018.01208. eCollection 2018. Front Pharmacol. 2018. PMID: 30405420 Free PMC article. Review. - Intrabody gene therapy ameliorates motor, cognitive, and neuropathological symptoms in multiple mouse models of Huntington's disease.
Southwell AL, Ko J, Patterson PH. Southwell AL, et al. J Neurosci. 2009 Oct 28;29(43):13589-602. doi: 10.1523/JNEUROSCI.4286-09.2009. J Neurosci. 2009. PMID: 19864571 Free PMC article.
References
- Colby, D. W., Garg, P., Holden, T., Chao, G., Webster, J. M., Messer, A., Ingram, V. M. & Wittrup, K. D. (2004) J. Mol. Biol. 342, 901–912. - PubMed
- Murphy, R. C. & Messer, A. (2004) Brain Res. Mol. Brain Res. 121, 141–145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials