Microevolution and history of the plague bacillus, Yersinia pestis - PubMed (original) (raw)
. 2004 Dec 21;101(51):17837-42.
doi: 10.1073/pnas.0408026101. Epub 2004 Dec 14.
Giovanna Morelli, Peixuan Zhu, Thierry Wirth, Ines Diehl, Barica Kusecek, Amy J Vogler, David M Wagner, Christopher J Allender, W Ryan Easterday, Viviane Chenal-Francisque, Patricia Worsham, Nicholas R Thomson, Julian Parkhill, Luther E Lindler, Elisabeth Carniel, Paul Keim
Affiliations
- PMID: 15598742
- PMCID: PMC535704
- DOI: 10.1073/pnas.0408026101
Microevolution and history of the plague bacillus, Yersinia pestis
Mark Achtman et al. Proc Natl Acad Sci U S A. 2004.
Abstract
The association of historical plague pandemics with Yersinia pestis remains controversial, partly because the evolutionary history of this largely monomorphic bacterium was unknown. The microevolution of Y. pestis was therefore investigated by three different multilocus molecular methods, targeting genomewide synonymous SNPs, variation in number of tandem repeats, and insertion of IS100 insertion elements. Eight populations were recognized by the three methods, and we propose an evolutionary tree for these populations, rooted on Yersinia pseudotuberculosis. The tree invokes microevolution over millennia, during which enzootic pestoides isolates evolved. This initial phase was followed by a binary split 6,500 years ago, which led to populations that are more frequently associated with human disease. These populations do not correspond directly to classical biovars that are based on phenotypic properties. Thus, we recommend that henceforth groupings should be based on molecular signatures. The age of Y. pestis inferred here is compatible with the dates of historical pandemic plague. However, it is premature to infer an association between any modern molecular grouping and a particular pandemic wave that occurred before the 20th century.
Figures
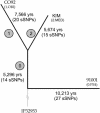
Fig. 1.
Age of Y. pestis. sSNPs were identified by pairwise genome comparisons between 91001 (0.PE4), CO92 (1.ORI), and KIM (2.MED). For each sSNP, one of the alternative nucleotides is present at the corresponding position within the genome of Y. pseudotuberculosis strain IP32953. sSNPs on branch 0 (Table 4) were identical in IP32953 and 91001 and also identical in KIM and CO92, but differed between these pairs. Other sSNPs were unique to the branches, as indicated. To calculate ages, the number of sSNPs was divided by the 777,520 potential sSNPs within the 3,250 homologous gene pairs, and that distance was then divided by the molecular clock rate of 3.4 × 10-9 per year.
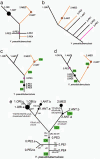
Fig. 2.
Evolutionary branch order within Y. pestis. (a_-d) Simplified branch order of the major groups as indicated by sSNPs (a), MLVA (b), and IS_100 insertions (c and d), based on data in Figs. 3, 6, and 7. The primary inconsistencies between a and b_-d are indicated in orange and purple. The differences in branch order between c and d reflect different interpretation of insertion events (green text). Nodes along branches are indicated by circles, the sizes of which indicate the number of isolates. (e) Consensus evolutionary order of IS_100 insertions (Yxx) and synonymous mutations (sxx). The diagram also indicates the inferred order of phenotypic changes (Rha-, Mel-, and Nit-) and nutritional mutations (glpD, napA316), except for the Nit- isolates in 2.ANT, which are not indicated. Sources of isolates according to grouping: 0.PE1, former Soviet Union (4 isolates); 0.PE2, former Soviet Union (3 isolates); 0.PE3, Africa (1 isolate); 0.PE4, China (1 isolate); 1.ANT, Africa (21 isolates); 1.ORI, global (95 isolates); 2.ANT, East Asia (5 isolates); and 2.MED, Kurdistan (26 isolates).
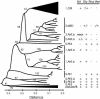
Fig. 3.
Relationships among 104 isolates according to MLVA. A neighbor-joining dendrogram was constructed from Hamming distances based on 43 variable number of tandem repeat loci. Individual isolates are shown except within 1.ORI (58 isolates) and pseudoTB (Y. pseudotuberculosis; 9 isolates), which were collapsed. Numbers within the dendrogram indicate high (>50%) bootstrap values associated with individual nodes. Group assignments according to sSNPs and the ability to reduce nitrate and ferment particular sugars (glycerol, rhamnose, and melibiose) are indicated at the right. For groups with mixed phenotypes, the predominant phenotype is indicated first. Exceptional strains were: 1.ORI Gly+, strain Nich51; 2.MED Gly- Mel+, pestoides J; and 2.ANT.a Nit-, Harbin 35, Nicholisk 41.
Fig. 4.
The napA613 mutation results in the inability to reduce nitrate. (A) Organization of the nap operon in Y. pestis. The only sequence differences between a 2.MED Nit- strain (IP564) and a 1.ANT Nit+ strain (IP554) within 5.9 kb spanning the nap operon was napA613, a stop codon. The predicted NapA protein from Y. pseudotuberculosis IP32953 differs by two other amino acids encoded by the nucleotides in bold type. (B) Complementation of nitrate reduction. Transformation of plasmid pBE696, containing the napA gene from IP32953, into 2.MED strains IP519 or IP616 (data not shown) restores their ability to reduce nitrate, as indicated by the red color of the growth medium.
Similar articles
- Genome sequence of the deep-rooted Yersinia pestis strain Angola reveals new insights into the evolution and pangenome of the plague bacterium.
Eppinger M, Worsham PL, Nikolich MP, Riley DR, Sebastian Y, Mou S, Achtman M, Lindler LE, Ravel J. Eppinger M, et al. J Bacteriol. 2010 Mar;192(6):1685-99. doi: 10.1128/JB.01518-09. Epub 2010 Jan 8. J Bacteriol. 2010. PMID: 20061468 Free PMC article. - Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis.
Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Achtman M, et al. Proc Natl Acad Sci U S A. 1999 Nov 23;96(24):14043-8. doi: 10.1073/pnas.96.24.14043. Proc Natl Acad Sci U S A. 1999. PMID: 10570195 Free PMC article. - Genome and Evolution of Yersinia pestis.
Cui Y, Song Y. Cui Y, et al. Adv Exp Med Biol. 2016;918:171-192. doi: 10.1007/978-94-024-0890-4_6. Adv Exp Med Biol. 2016. PMID: 27722863 Review. - Pseudogene accumulation might promote the adaptive microevolution of Yersinia pestis.
Tong Z, Zhou D, Song Y, Zhang L, Pei D, Han Y, Pang X, Li M, Cui B, Wang J, Guo Z, Qi Z, Jin L, Zhai J, Du Z, Wang J, Wang X, Yu J, Wang J, Huang P, Yang H, Yang R. Tong Z, et al. J Med Microbiol. 2005 Mar;54(Pt 3):259-268. doi: 10.1099/jmm.0.45752-0. J Med Microbiol. 2005. PMID: 15713609 - Review of genotyping methods for Yersinia pestis in Madagascar.
Randriantseheno LN, Andrianaivoarimanana V, Pizarro-Cerdá J, Wagner DM, Rajerison M. Randriantseheno LN, et al. PLoS Negl Trop Dis. 2024 Jun 27;18(6):e0012252. doi: 10.1371/journal.pntd.0012252. eCollection 2024 Jun. PLoS Negl Trop Dis. 2024. PMID: 38935608 Free PMC article. Review.
Cited by
- Insights from genomic comparisons of genetically monomorphic bacterial pathogens.
Achtman M. Achtman M. Philos Trans R Soc Lond B Biol Sci. 2012 Mar 19;367(1590):860-7. doi: 10.1098/rstb.2011.0303. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22312053 Free PMC article. Review. - Combinational deletion of three membrane protein-encoding genes highly attenuates yersinia pestis while retaining immunogenicity in a mouse model of pneumonic plague.
Tiner BL, Sha J, Kirtley ML, Erova TE, Popov VL, Baze WB, van Lier CJ, Ponnusamy D, Andersson JA, Motin VL, Chauhan S, Chopra AK. Tiner BL, et al. Infect Immun. 2015 Apr;83(4):1318-38. doi: 10.1128/IAI.02778-14. Epub 2015 Jan 20. Infect Immun. 2015. PMID: 25605764 Free PMC article. - Single-Nucleotide Polymorphisms Reveal Spatial Diversity Among Clones of Yersinia pestis During Plague Outbreaks in Colorado and the Western United States.
Lowell JL, Antolin MF, Andersen GL, Hu P, Stokowski RP, Gage KL. Lowell JL, et al. Vector Borne Zoonotic Dis. 2015 May;15(5):291-302. doi: 10.1089/vbz.2014.1714. Vector Borne Zoonotic Dis. 2015. PMID: 25988438 Free PMC article. - Genome sequence of the deep-rooted Yersinia pestis strain Angola reveals new insights into the evolution and pangenome of the plague bacterium.
Eppinger M, Worsham PL, Nikolich MP, Riley DR, Sebastian Y, Mou S, Achtman M, Lindler LE, Ravel J. Eppinger M, et al. J Bacteriol. 2010 Mar;192(6):1685-99. doi: 10.1128/JB.01518-09. Epub 2010 Jan 8. J Bacteriol. 2010. PMID: 20061468 Free PMC article. - Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer.
Pearson T, Giffard P, Beckstrom-Sternberg S, Auerbach R, Hornstra H, Tuanyok A, Price EP, Glass MB, Leadem B, Beckstrom-Sternberg JS, Allan GJ, Foster JT, Wagner DM, Okinaka RT, Sim SH, Pearson O, Wu Z, Chang J, Kaul R, Hoffmaster AR, Brettin TS, Robison RA, Mayo M, Gee JE, Tan P, Currie BJ, Keim P. Pearson T, et al. BMC Biol. 2009 Nov 18;7:78. doi: 10.1186/1741-7007-7-78. BMC Biol. 2009. PMID: 19922616 Free PMC article.
References
- Yersin, A. (1894) Ann. Inst. Pasteur 2, 428-430.
- Radnedge, L., Agron, P. G., Worsham, P. L. & Andersen, G. L. (2002) Microbiology 148, 1687-1698. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical