A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase - PubMed (original) (raw)
A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase
Kazufumi Mochizuki et al. Genes Dev. 2005.
Abstract
Previous studies indicated that genome rearrangement involving DNA sequence elimination that occurs at late stages of conjugation in Tetrahymena is epigenetically controlled by siRNA-like scan (scn) RNAs produced from nongenic, heterogeneous, bidirectional, micronuclear transcripts synthesized at early stages of conjugation. Here, we show that Dcl1p, one of three Tetrahymena Dicer-like enzymes, is required for processing the micronuclear transcripts to scnRNAs. DCL1 is also required for methylation of histone H3 at Lys 9, which, in wild-type cells, specifically occurs on the sequences (IESs) being eliminated. These results argue that Dcl1p processes nongenic micronuclear transcripts to scnRNAs and is required for IES elimination. This is the first evidence linking nongenic micronuclear transcripts, scnRNAs, and genome rearrangement. Dcl1p also is required for proper mitotic and meiotic segregation of micronuclear chromosomes and for normal chromosome alignment in meiotic prophase, suggesting that DCL1 has multiple functions in regulating chromosome dynamics.
Figures
Figure 1.
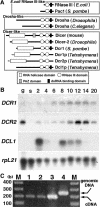
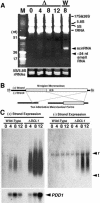
Characterization of _dicer_-related genes. (A) Comparison of RNase III enzymes. Three classes of RNase III proteins, Escherichia coli RNase III-like, Drosha-like, and Dicer-like are schematically drawn. N termini of Dcr1p and Dcr2p (dotted lines) have not been determined experimentally. (B) mRNA expression of dicer-related genes analyzed on Northern blot. (g) Log-phase growing cells; (s) starved cells; (2–20) conjugating cells (numbers indicate hours post-mixing). Gene-specific probes were used for hybridization. Hybridization to a probe for rpL21, encoding a ribosomal protein was used as a loading control. (C) DCL1 mRNA is expressed in vegetative cells. Total RNA extracted from log-phase growing wild-type B2086 (lane 1), CU428 (lane 2), and mating B2086 and CU428 cells at 3 h post-mixing (lane 3) were used for RT–PCR. (Lane 4) Total genomic DNA of CU428 was also used for PCR. A 55-b intron is present in the genomic region, enabling the PCR products from genomic DNA (246 b) and cDNA (191 b) to be distinguished.
Figure 4.
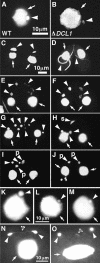
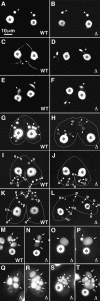
DCL1 is required for Mic chromosome maintenance. (A,B) Comparison of wild-type (A) and DCL1 knockout (B) cells. The log-phase growing cells were fixed and stained with DAPI. The Mics in DCL1 knockout cells contain much less DNA than those of wild-type cells. A and B share the same scale bar shown in A.(C–J) The Mics of DCL1 knockout cells cannot transmit genetic material to the next generation. Wild-type cells and DCL1 knockout cells were crossed, and the fixed cells were stained with DAPI. Mic abnormalities suggest that wild-type and DCL1 knockout cells were at left and right, respectively. Cells were in stages of early crescent (meiotic prophase; Stage II) (C), late crescent (meiotic prophase; Stage IV) (D), first meiotic division completed (E,F), second meiotic division completed (G), prezygotic mitosis (H), pronuclear (unilateral) exchange (I), and pronuclear (unilateral) exchange completed (J). For stages, see Sugai and Hiwatashi (1974) and Cole et al. (1997). The Macs and the Mics are marked with arrows and arrowheads, respectively. In H, the selected, mitotically dividing haploid Mic is marked with an s, and in I and J, pronuclei are marked with a p. C–J share the same scale bar shown in C. (K,L) A DCL1 knockout RI strain loses Mic chromosomes during vegetative growth. DCL1 knockout RI cells (Δ_DCL1_-14A-RI) were fixed at about 12 (K) and 60 (L) fissions after Round I genomic exclusion and stained with DAPI. (M) A DCL1 somatic knockout strain loses Mic chromosomes. DCL1 somatic knockout cells in interphase were fixed and stained with DAPI. K–M share the same scale bar shown in L. (N,O) DCL1 knockout cells show lagging chromosomes during Mic mitosis. DCL1 knockout RI cells in late stages of Mic mitosis were fixed and stained with DAPI. Two daughter Mics were often connected by DNA (N, arrowhead with asterisk); what appear to be chromosomes or chromosome fragments were often left between the daughter Mics (O, arrowhead with asterisk). N and O share the same 10-μm scale bar shown in N. In all pictures, the Macs and the Mics are marked with arrows and arrowheads, respectively.
Figure 2.
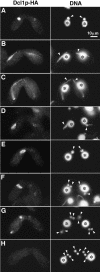
Localization of Dcl1p-HA. The _DCL1_-HA strain was mated with the wild-type B2086 strain and processed for indirect immunofluorescent staining. Dcl1p-HA was localized by anti-HA antibody (left) and DNA was stained with DAPI (right). Early crescent (A), mid-crescent (B), and late-crescent (C) stages of meiotic prophase. (D) Chromosome condensation. (E,F) First meiosis. (G) Second meiosis. (H) Second meiosis completed. All photos share a common scale bar shown in A. Arrows and asterisks indicate Mics and Macs, respectively. For stages of mating, see Cole et al. (1997).
Figure 3.
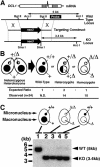
DCL1 is not essential for vegetative growth. (A) Schematic drawings of the DCL1 locus and the knockout construct used to disrupt it. The N-terminal half of the DCL1 coding sequence was replaced by the neo3 cassette that confers paromomycin resistance (pm-r) in Tetrahymena. The knockout construct was introduced into the DCL1 locus by homologous recombination. (B) Viability of homozygous DCL1 KO cells. Two heterozygous DCL1 knockout heterokaryons were mated, and their progeny were selected using pm. The genotypes of the pm-r progeny were analyzed. Of 24 pm-r progeny analyzed, 10 were homozygous. Because wild-type progeny are pm-sensitive and could not be distinguished from the parental strains, their fraction of the progeny was not determined. (C) Complete replacement of endogenous DCL1 loci in the knockout strains. Total DNA isolated from wild-type CU428 (lane 1), DCL1 knockout homozygous homokaryons (lanes 2,3), and DCL1 somatic knockout strains (lanes 4,5) was digested with AccI and SmaI and the Southern blot was hybridized with the probe shown in A. Positions of the bands for wild-type (WT) and disrupted (KO) loci are indicated with arrowheads. The faint, wild-type size bands observed in DCL1 somatic knockout strains were from Mics that had wild-type DCL1 loci.
Figure 5.
DCL1 knockout cells have defects in Mic chromosome dynamics during conjugation. Two wild-type strains (WT) or two Round I DCL1 knockout strains (Δ) were mated, fixed, and stained with DAPI. (A,B) Immediately after pairing. (C,D) Meiotic prophase (crescent stage). (E,F) Chromosome condensation. (G,H) After the second meiotic division. (I,J) Nuclear selection. (K,L) Pronuclear exchange. (M–T) Nuclear alignment stage. (O) Only a new Mac developed. (P) A Mac and a Mic developed. (Q) Four Macs developed. (R) Six Macs and two (one is not visible) Mics developed. (S) Three (incomplete) Macs and five Mics developed. (T) Four Macs and six Mics developed. (Asterisks) (parental) Macs; (arrowheads) Mics; (arrows) developing new Macs; (s) selected haploid Mics; (u) unselected, eliminating Mics; (p) pronuclei. In G–L, outlines of cells were traced. For stages, see Cole et al. (1997). All pictures share the same scale bar shown in A.
Figure 6.
Expression of scnRNA and nongenic Mic transcripts. (A) Total RNA was extracted from mating DCL1 knockout RI cells (Δ) at 0, 4, 8, and 12 h post-mixing or from wild-type cells (W) at 8 h post-mixing. RNA from 5 × 104 cells was analyzed in 12% acrylamide-urea gel and stained with ethidium bromide. In vitro-transcribed 17, 26, and 51 nt RNAs were used as markers (M). The gel was partially destained, and the amounts of 5S and 5.8S rRNAs are shown as loading controls. (B) Schematic drawing of the Mic M-region sequence (top) and its two alternatively processed Mac forms (bottom). (Open box) IES; (gray box) IES eliminated only in the long-deletion form; (horizontal bars) Mac-destined sequences (MDSs). Promoters for T3 and T7 RNA polymerases were added at the 5′ and 3′ ends of Mic M-region sequence, respectively. (C) Northern blot made with total RNA extracted from equal numbers of wild-type and DCL1 knockout RI cells at 0, 4, 8, and 12 h post-mixing and hybridized with probes complementary to (+) strand (left) and (–) strand (right) of the Mic M-region made by in vitro transcription with T3 and T7 RNA polymerase, respectively. Positions for 17S + 28S rRNAs and tRNAs are marked with r and t, respectively. The blot used for (+) strand probe was reprobed with a PDD1 probe as a marker for conjugation (left, bottom). Increased expression of PDD1 in DCL1 knockout cells at late stages of conjugation (12 h post-mixing) probably reflected arrested conjugation in the late stages.
Figure 7.
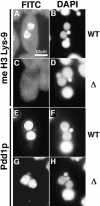
Expression of methylated H3 Lys 9 and Pdd1p. (A–D) Localization of methylated H3 Lys 9. Wild-type (A,B) or DCL1 knockout RI strain (C,D) at nuclear alignment stage (12 h post-mixing) were stained with anti-methylated H3 Lys 9 antibody (A,C) or with DAPI (B,D). (E–H) Localization of Pdd1p. Wild-type (E,F) or DCL1 knockout RI strain (G,H) at nuclear alignment stage were stained with anti-Pdd1p antibody (E,G) or with DAPI (F,H). All photos share the scale bar shown in A.
Similar articles
- Programmed DNA elimination in Tetrahymena: a small RNA-mediated genome surveillance mechanism.
Kataoka K, Mochizuki K. Kataoka K, et al. Adv Exp Med Biol. 2011;722:156-73. doi: 10.1007/978-1-4614-0332-6_10. Adv Exp Med Biol. 2011. PMID: 21915788 Free PMC article. Review. - Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila.
Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL. Malone CD, et al. Mol Cell Biol. 2005 Oct;25(20):9151-64. doi: 10.1128/MCB.25.20.9151-9164.2005. Mol Cell Biol. 2005. PMID: 16199890 Free PMC article. - Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena.
Liu Y, Mochizuki K, Gorovsky MA. Liu Y, et al. Proc Natl Acad Sci U S A. 2004 Feb 10;101(6):1679-84. doi: 10.1073/pnas.0305421101. Epub 2004 Jan 30. Proc Natl Acad Sci U S A. 2004. PMID: 14755052 Free PMC article. - Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena.
Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K. Aronica L, et al. Genes Dev. 2008 Aug 15;22(16):2228-41. doi: 10.1101/gad.481908. Genes Dev. 2008. PMID: 18708581 Free PMC article. - Small RNAs in genome rearrangement in Tetrahymena.
Mochizuki K, Gorovsky MA. Mochizuki K, et al. Curr Opin Genet Dev. 2004 Apr;14(2):181-7. doi: 10.1016/j.gde.2004.01.004. Curr Opin Genet Dev. 2004. PMID: 15196465 Review.
Cited by
- Genome-Scale Analysis of Programmed DNA Elimination Sites in Tetrahymena thermophila.
Fass JN, Joshi NA, Couvillion MT, Bowen J, Gorovsky MA, Hamilton EP, Orias E, Hong K, Coyne RS, Eisen JA, Chalker DL, Lin D, Collins K. Fass JN, et al. G3 (Bethesda). 2011 Nov;1(6):515-22. doi: 10.1534/g3.111.000927. Epub 2011 Nov 1. G3 (Bethesda). 2011. PMID: 22384362 Free PMC article. - RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena.
Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. Liu Y, et al. Genes Dev. 2007 Jun 15;21(12):1530-45. doi: 10.1101/gad.1544207. Genes Dev. 2007. PMID: 17575054 Free PMC article. - Subtraction by addition: domesticated transposases in programmed DNA elimination.
Motl JA, Chalker DL. Motl JA, et al. Genes Dev. 2009 Nov 1;23(21):2455-60. doi: 10.1101/gad.1864609. Genes Dev. 2009. PMID: 19884252 Free PMC article. - Chromatin-associated ncRNA activities.
Keller C, Bühler M. Keller C, et al. Chromosome Res. 2013 Dec;21(6-7):627-41. doi: 10.1007/s10577-013-9390-8. Chromosome Res. 2013. PMID: 24249576 Free PMC article. Review. - Programmed DNA elimination in Tetrahymena: a small RNA-mediated genome surveillance mechanism.
Kataoka K, Mochizuki K. Kataoka K, et al. Adv Exp Med Biol. 2011;722:156-73. doi: 10.1007/978-1-4614-0332-6_10. Adv Exp Med Biol. 2011. PMID: 21915788 Free PMC article. Review.
References
- Allis C.D., Richman, R., Gorovsky, M.A., Ziegler, Y.S., Touchstone, B., Bradley, W.A., and Cook, R.G. 1986. hv1 is an evolutionarily conserved H2A variant that is preferentially associated with active genes. J. Biol. Chem. 261: 1941–1948. - PubMed
- Carmell M.A. and Hannon, G.J. 2004. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 11: 214–218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources