GenomeViz: visualizing microbial genomes - PubMed (original) (raw)
Comparative Study
GenomeViz: visualizing microbial genomes
Rohit Ghai et al. BMC Bioinformatics. 2004.
Abstract
Background: An increasing number of microbial genomes are being sequenced and deposited in public databases. In addition, several closely related strains are also being sequenced in order to understand the genetic basis of diversity and mechanisms that lead to the acquisition of new genetic traits. These exercises have necessitated the requirement for visualizing microbial genomes and performing genome comparisons on a finer scale. We have developed GenomeViz to enable rapid visualization and subsequent comparisons of several microbial genomes in an interactive environment.
Results: Here we describe a program that allows visualization of both qualitative and quantitative information from complete and partially sequenced microbial genomes. Using GenomeViz, data deriving from studies on genomic islands, gene/protein classifications, GC content, GC skew, whole genome alignments, microarrays and proteomics may be plotted. Several genomes can be visualized interactively at the same time from a comparative genomic perspective and publication quality circular genome plots can be created.
Conclusions: GenomeViz should allow researchers to perform visualization and comparative analysis of up to eight different microbial genomes simultaneously.
Figures
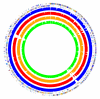
Figure 1
Whole genome alignments of five Listeria strains/species. From outside to inside: L. monocytogenes EGDe serovar 1/2a COG categories (outer two circles), L. monocytogenes F6854 serovar 1/2a (blue, 133 contigs), L. monocytogenes F2365 serovar 4b (red, whole genome), L. monocytogenes H7858 serovar 4b (orange, 180 contigs) and L. innocua (innermost, green, whole genome). All genomes were aligned separately to L. monocytogenes EGDe with AVID. Sequence data for strains L. monocytogenes strains F6854 and H7858 was obtained from The Institute for Genomic Research [17].
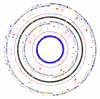
Figure 2
A typical image generated by GenomeViz. From outside to inside: Listeria monocytogenes COG categories (two circles), horizontally transferred gene categories in L. monocytogenes identified using SIGI (two circles), mean centered GC% of L. monocytogenes genes (red-above mean, blue-below mean, one circle), GC% gradient (red-high GC%, green-low GC%, one circle), Listeria innocua COG categories (two circles), horizontally transferred gene categories in L. innocua identified using SIGI (two circles), mean centered GC% of L. innocua genes (red-above mean, blue-below mean, one circle), GC% of L. innocua genes shown as a line graph (innermost circle). When created in GenomeViz, this image is fully interactive and any plotted circle may be queried. It also shows the different ways in which qualitative or numerical data may be plotted. Differences in the horizontally transferred genes in the two Listeria species may be examined and related to GC content in the region.
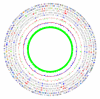
Figure 3
**Comparing data from different sources using GenomeViz.**The figure shows a comparison of the distribution of horizontally transferred genes in Escherichia coli K12 compiled from three different sources. From outside to inside: Escherichia coli K12 COG categories (two circles), genes identified by SIGI (two circles), genes listed in the Horizontal Gene Transfer Database [18] (two circles), standard deviations of genes identified by IslandPath (single circle, red +ve, green -ve), mean centered GC content of the genome (red: above mean, blue: below mean), GC content of the genome again as a single-sided line plot (green).
Similar articles
- CGAT: a comparative genome analysis tool for visualizing alignments in the analysis of complex evolutionary changes between closely related genomes.
Uchiyama I, Higuchi T, Kobayashi I. Uchiyama I, et al. BMC Bioinformatics. 2006 Oct 24;7:472. doi: 10.1186/1471-2105-7-472. BMC Bioinformatics. 2006. PMID: 17062155 Free PMC article. - The genome BLASTatlas-a GeneWiz extension for visualization of whole-genome homology.
Hallin PF, Binnewies TT, Ussery DW. Hallin PF, et al. Mol Biosyst. 2008 May;4(5):363-71. doi: 10.1039/b717118h. Epub 2008 Mar 17. Mol Biosyst. 2008. PMID: 18414733 - ArrayOme: a program for estimating the sizes of microarray-visualized bacterial genomes.
Ou HY, Smith R, Lucchini S, Hinton J, Chaudhuri RR, Pallen M, Barer MR, Rajakumar K. Ou HY, et al. Nucleic Acids Res. 2005 Jan 7;33(1):e3. doi: 10.1093/nar/gni005. Nucleic Acids Res. 2005. PMID: 15640440 Free PMC article. - Visualization of sequence and structural features of genomes and chromosome fragments. Application to CpG islands, Alu sequences and whole genomes.
Subirana JA, Anokian E. Subirana JA, et al. Gene. 2011 Mar 1;473(2):76-81. doi: 10.1016/j.gene.2010.11.008. Epub 2010 Dec 16. Gene. 2011. PMID: 21167919 Review. - How do we compare hundreds of bacterial genomes?
Field D, Wilson G, van der Gast C. Field D, et al. Curr Opin Microbiol. 2006 Oct;9(5):499-504. doi: 10.1016/j.mib.2006.08.008. Epub 2006 Aug 30. Curr Opin Microbiol. 2006. PMID: 16942900 Review.
Cited by
- Safety assessment of Bifidobacterium longum JDM301 based on complete genome sequences.
Wei YX, Zhang ZY, Liu C, Malakar PK, Guo XK. Wei YX, et al. World J Gastroenterol. 2012 Feb 7;18(5):479-88. doi: 10.3748/wjg.v18.i5.479. World J Gastroenterol. 2012. PMID: 22346255 Free PMC article. - The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages.
Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, Hain T. Mraheil MA, et al. Nucleic Acids Res. 2011 May;39(10):4235-48. doi: 10.1093/nar/gkr033. Epub 2011 Jan 29. Nucleic Acids Res. 2011. PMID: 21278422 Free PMC article. - A systematic study of the whole genome sequence of Amycolatopsis methanolica strain 239T provides an insight into its physiological and taxonomic properties which correlate with its position in the genus.
Tang B, Xie F, Zhao W, Wang J, Dai S, Zheng H, Ding X, Cen X, Liu H, Yu Y, Zhou H, Zhou Y, Zhang L, Goodfellow M, Zhao GP. Tang B, et al. Synth Syst Biotechnol. 2016 Sep 1;1(3):169-186. doi: 10.1016/j.synbio.2016.05.001. eCollection 2016 Sep. Synth Syst Biotechnol. 2016. PMID: 29062941 Free PMC article. - Genomorama: genome visualization and analysis.
Gans JD, Wolinsky M. Gans JD, et al. BMC Bioinformatics. 2007 Jun 14;8:204. doi: 10.1186/1471-2105-8-204. BMC Bioinformatics. 2007. PMID: 17570856 Free PMC article. - Whole-genome sequence of Listeria welshimeri reveals common steps in genome reduction with Listeria innocua as compared to Listeria monocytogenes.
Hain T, Steinweg C, Kuenne CT, Billion A, Ghai R, Chatterjee SS, Domann E, Kärst U, Goesmann A, Bekel T, Bartels D, Kaiser O, Meyer F, Pühler A, Weisshaar B, Wehland J, Liang C, Dandekar T, Lampidis R, Kreft J, Goebel W, Chakraborty T. Hain T, et al. J Bacteriol. 2006 Nov;188(21):7405-15. doi: 10.1128/JB.00758-06. Epub 2006 Aug 25. J Bacteriol. 2006. PMID: 16936040 Free PMC article.
References
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous