HIV-1 encoded candidate micro-RNAs and their cellular targets - PubMed (original) (raw)
Comparative Study
HIV-1 encoded candidate micro-RNAs and their cellular targets
Yamina Bennasser et al. Retrovirology. 2004.
Abstract
MicroRNAs (miRNAs) are small RNAs of 21-25 nucleotides that specifically regulate cellular gene expression at the post-transcriptional level. miRNAs are derived from the maturation by cellular RNases III of imperfect stem loop structures of ~ 70 nucleotides. Evidence for hundreds of miRNAs and their corresponding targets has been reported in the literature for plants, insects, invertebrate animals, and mammals. While not all of these miRNA/target pairs have been functionally verified, some clearly serve roles in regulating normal development and physiology. Recently, it has been queried whether the genome of human viruses like their cellular counterpart also encode miRNA. To date, there has been only one report pertaining to this question. The Epstein-Barr virus (EBV) has been shown to encode five miRNAs. Here, we extend the analysis of miRNA-encoding potential to the human immunodeficiency virus (HIV). Using computer-directed analyses, we found that HIV putatively encodes five candidate pre-miRNAs. We then matched deduced mature miRNA sequences from these 5 pre-miRNAs against a database of 3' untranslated sequences (UTR) from the human genome. These searches revealed a large number of cellular transcripts that could potentially be targeted by these viral miRNA (vmiRNA) sequences. We propose that HIV has evolved to use vmiRNAs as a means to regulate cellular milieu for its benefit.
Figures
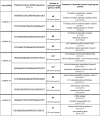
Figure 1
Sequences and localization of HIV-encoded miRNA candidates. a) Locations for 5 predicted pre-miRNAs candidates in the pNL4-3 genome are shown. b) The folded structures of the 5 viral pre-miRNAs from pNL4-3 (Accession Number AF 324493) [17] are illustrated. Folded pre-miRNAs and their corresponding predicted mature viral miRNA (red) are listed. Nucleotide positions (where 1 is the initiation of transcription) in the pNL4-3 genome are presented in the right column.
Figure 2
Potential cellular targets for each of the vmiRNAs. The two deduced mature vmiRNAs predicted from each precursor miRNA are shown. The mature vmiRNA sequences were individually searched against a database of human 3'UTRs using imperfect complementarity criteria as described in the text. The number of potential candidate cellular RNA targets is enumerated. Most of the cellular targets are incompletely characterized expressed sequence tag (EST) clones, with a subset of targets being known genes. For each predicted vmiRNA, we list two examples of known cellular gene targets at the right. A full list of targets is available upon request.
Similar articles
- HBV-encoded microRNA candidate and its target.
Jin WB, Wu FL, Kong D, Guo AG. Jin WB, et al. Comput Biol Chem. 2007 Apr;31(2):124-6. doi: 10.1016/j.compbiolchem.2007.01.005. Epub 2007 Jan 26. Comput Biol Chem. 2007. PMID: 17350341 - The HIV-2 TAR RNA domain as a potential source of viral-encoded miRNA. A reconnaissance study.
Purzycka KJ, Adamiak RW. Purzycka KJ, et al. Nucleic Acids Symp Ser (Oxf). 2008;(52):511-2. doi: 10.1093/nass/nrn259. Nucleic Acids Symp Ser (Oxf). 2008. PMID: 18776478 - Targets for human encoded microRNAs in HIV genes.
Hariharan M, Scaria V, Pillai B, Brahmachari SK. Hariharan M, et al. Biochem Biophys Res Commun. 2005 Dec 2;337(4):1214-8. doi: 10.1016/j.bbrc.2005.09.183. Epub 2005 Oct 7. Biochem Biophys Res Commun. 2005. PMID: 16236258 - Prediction of microRNA targets.
Mazière P, Enright AJ. Mazière P, et al. Drug Discov Today. 2007 Jun;12(11-12):452-8. doi: 10.1016/j.drudis.2007.04.002. Epub 2007 Apr 26. Drug Discov Today. 2007. PMID: 17532529 Review. - siRNA, miRNA and HIV: promises and challenges.
Yeung ML, Bennasser Y, LE SY, Jeang KT. Yeung ML, et al. Cell Res. 2005 Nov-Dec;15(11-12):935-46. doi: 10.1038/sj.cr.7290371. Cell Res. 2005. PMID: 16354572 Review.
Cited by
- Cotranscriptional Chromatin Remodeling by Small RNA Species: An HTLV-1 Perspective.
Aliya N, Rahman S, Khan ZK, Jain P. Aliya N, et al. Leuk Res Treatment. 2012;2012:984754. doi: 10.1155/2012/984754. Epub 2012 Feb 9. Leuk Res Treatment. 2012. PMID: 23213554 Free PMC article. - NMR Studies of Retroviral Genome Packaging.
Boyd PS, Brown JB, Brown JD, Catazaro J, Chaudry I, Ding P, Dong X, Marchant J, O'Hern CT, Singh K, Swanson C, Summers MF, Yasin S. Boyd PS, et al. Viruses. 2020 Sep 30;12(10):1115. doi: 10.3390/v12101115. Viruses. 2020. PMID: 33008123 Free PMC article. Review. - Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus.
Samols MA, Hu J, Skalsky RL, Renne R. Samols MA, et al. J Virol. 2005 Jul;79(14):9301-5. doi: 10.1128/JVI.79.14.9301-9305.2005. J Virol. 2005. PMID: 15994824 Free PMC article. - Ebola Virus Encodes Two microRNAs in Huh7-Infected Cells.
Diallo I, Husseini Z, Guellal S, Vion E, Ho J, Kozak RA, Kobinger GP, Provost P. Diallo I, et al. Int J Mol Sci. 2022 May 7;23(9):5228. doi: 10.3390/ijms23095228. Int J Mol Sci. 2022. PMID: 35563619 Free PMC article. - Establishment of a miRNA profile in paediatric HIV-1 patients and its potential as a biomarker for effectiveness of the combined antiretroviral therapy.
Consuegra I, Gasco S, Serramía MJ, Jiménez JL, Mellado MJ, Muñoz-Fernández MÁ. Consuegra I, et al. Sci Rep. 2021 Dec 6;11(1):23477. doi: 10.1038/s41598-021-03020-5. Sci Rep. 2021. PMID: 34873266 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources