Polybromo protein BAF180 functions in mammalian cardiac chamber maturation - PubMed (original) (raw)
Comparative Study
. 2004 Dec 15;18(24):3106-16.
doi: 10.1101/gad.1238104.
Affiliations
- PMID: 15601824
- PMCID: PMC535920
- DOI: 10.1101/gad.1238104
Comparative Study
Polybromo protein BAF180 functions in mammalian cardiac chamber maturation
Zhong Wang et al. Genes Dev. 2004.
Abstract
BAF and PBAF are two related mammalian chromatin remodeling complexes essential for gene expression and development. PBAF, but not BAF, is able to potentiate transcriptional activation in vitro mediated by nuclear receptors, such as RXRalpha, VDR, and PPARgamma. Here we show that the ablation of PBAF-specific subunit BAF180 in mouse embryos results in severe hypoplastic ventricle development and trophoblast placental defects, similar to those found in mice lacking RXRalpha and PPARgamma. Embryonic aggregation analyses reveal that in contrast to PPARgamma-deficient mice, the heart defects are likely a direct result of BAF180 ablation, rather than an indirect consequence of trophoblast placental defects. We identified potential target genes for BAF180 in heart development, such as S100A13 as well as retinoic acid (RA)-induced targets RARbeta2 and CRABPII. Importantly, BAF180 is recruited to the promoter of these target genes and BAF180 deficiency affects the RA response for CRABPII and RARbeta2. These studies reveal unique functions of PBAF in cardiac chamber maturation.
Figures
Figure 1.
Generation of BAF180-deficient mice. (A) KO scheme applied for generation of a BAF180 null allele in ES cells. Wild-type genomic structure (WT, top line) is replaced by a neo-resistant knockout allele (KO, neor, bottom line) through homologous recombination within the grey fragments. Cre-mediated recombination between the two loxP sites converts the KO allele to neo-sensitive and lacZ negative (neos). (B) For BAF180 KO Southern analysis, Spe I digestion produces a 7.8-kb fragment for wild type (WT) allele and a 5.9-kb fragment for a mutant allele. The probe used is indicated by a small black box located between the Spe I sites and distal to the 5′ recombination region. (C) PCR-based genotyping analyses of the progenies of BAF180 heterozygous crosses. (D) Western blot indicated that the mouse BAF180 protein is absent in cells derived from homozygous BAF180 KO mice. TAFII250 served as a positive control.
Figure 2.
BAF180 expression in mouse tissues. (A) X-Gal staining of β-galactosidase activity revealed that BAF180 is expressed ubiquitously at E11.5 embryo. (B) In situ hybridization of E14.5 placenta with a BAF180-specific antisense RNA probe. (LB) Labyrinthine; (SP) spongiotrophoblast.
Figure 3.
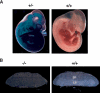
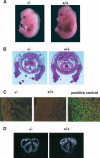
Cardiac defects in BAF180-deficient embryos. (A) Wild-type (WT) and null BAF180 embryos at E14.5. Note the edema along the back of the mutant embryo. (B) Transverse sections of a wild-type (WT) and a mutant E14.5 heart. The mutant heart has severe hypoplastic myocardial wall and ventricular septal defects. (C) Tunnel assays indicated that neither BAF180 wild-type (WT) nor null E14.5 heart tissues showed any detectable cell death. A DNase I-treated heart tissue section was used as a positive control. (D) In situ hybridization of E14.5 hearts indicated that the expression pattern of MLC-2a is largely normal in a BAF180 null heart. (E) Edema; (VS) ventricular septum; (VSD) ventricular septal defect.
Figure 4.
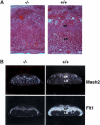
Placental defects in BAF180-deficient embryos. (A) Compared with wild type (WT) at E13.5, placentas from BAF180 null fetuses have indistinct spongiotrophoblast and labyrinthine layers. (B) In situ hybridization of E14.5 placentas with placental markers Mash2 and Flt1. (LB) Labyrinthine; (SP) spongiotrophoblast.
Figure 5.
Wild-type tetraploid embryo rescue did not restore normal heart development in BAF180 null embryos. (A) Contribution of BAF180 wild-type (WT) alleles in chimeras derived from aggregation between wild-type tetraploid and BAF180 mutant diploid embryos is analyzed by PCR. Lanes 1 and 2 identified two embryos derived from BAF180 nulls aggregated with wild-type tetraploid embryos. Lanes 3 and 4 identified two controls from normal intercross of BAF180 Hets. (B) A chimeric placenta derived from aggregation of a null BAF180 embryo with two wild-type (WT) tetraploid embryos appears to be normal. (C) Tetraploid aggregation could not rescue the heart defects. Histological analysis of the heart from the embryo identified in A, lane 1. All 10 null embryos identified from 42 recovered chimeras showed very similar phenotypes. (E) Edema; (LB) labyrinthine; (SP) spongiotrophoblast; (VSD) ventricular septal defect.
Figure 6.
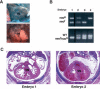
Wild-type ES cell rescue restored the normal heart development of BAF180 null embryos. (A) X-Gal staining demonstrated variable chimeric contribution of wild-type (WT) ES cells in an embryonic heart derived from aggregation between wild-type ES cells and BAF180 mutant diploid embryos. Less chimeric X-Gal staining represented more ES contribution. (B) PCR analysis demonstrated contribution of BAF180 wild-type (WT) alleles in chimeras derived from aggregation between wild-type ES cells and mutant diploid embryos. PCR was performed with DNA samples obtained from embryonic limb, tail, brain, yolk sac, and placenta (only results from brain tissues were illustrated). Embryo 1 is a null embryo with little wild-type ES contribution, while embryo 2 is a null embryo with substantial ES contribution. Embryos 3 and 4 are two Het embryos aggregated with wild-type ES cells. (C) Contribution of wild-type (WT) ES cells into BAF180 null embryos rescued the heart defects. Histological analysis of the hearts from the two null embryos identified in B. (H) Heart; (VS) ventricular septum.
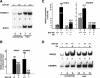
Figure 7.
Identification of BAF180 target genes. (A) ChIP assays show binding of BAF180 to promoter regions of S100A13 but not Grb10. (B) Quantitative RT-PCR analysis show decreased expression of both RARβ2 and CRABPII in BAF180 null embryonic hearts. (C) Quantitative RT-PCR analysis of RARβ2 and CRABPII expression in ES cells. Loss of BAF180 led to decreased expression of these two genes, and abolished the RA response for CRABPII and decreased RA response for RARβ2. (D) ChIP assays show recruitment of BAF180 to promoter regions of RARβ2 and CRABPII.
Similar articles
- PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes.
Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K, Wang W. Yan Z, et al. Genes Dev. 2005 Jul 15;19(14):1662-7. doi: 10.1101/gad.1323805. Epub 2005 Jun 28. Genes Dev. 2005. PMID: 15985610 Free PMC article. - IL-10 transcription is negatively regulated by BAF180, a component of the SWI/SNF chromatin remodeling enzyme.
Wurster AL, Precht P, Becker KG, Wood WH 3rd, Zhang Y, Wang Z, Pazin MJ. Wurster AL, et al. BMC Immunol. 2012 Feb 15;13:9. doi: 10.1186/1471-2172-13-9. BMC Immunol. 2012. PMID: 22336179 Free PMC article. - Baf60c is essential for function of BAF chromatin remodelling complexes in heart development.
Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Lickert H, et al. Nature. 2004 Nov 4;432(7013):107-12. doi: 10.1038/nature03071. Nature. 2004. PMID: 15525990 - BAF180: Its Roles in DNA Repair and Consequences in Cancer.
Hopson S, Thompson MJ. Hopson S, et al. ACS Chem Biol. 2017 Oct 20;12(10):2482-2490. doi: 10.1021/acschembio.7b00541. Epub 2017 Sep 28. ACS Chem Biol. 2017. PMID: 28921948 Review. - Cellular Retinoic Acid Binding Proteins: Genomic and Non-genomic Functions and their Regulation.
Wei LN. Wei LN. Subcell Biochem. 2016;81:163-178. doi: 10.1007/978-94-024-0945-1_6. Subcell Biochem. 2016. PMID: 27830504 Review.
Cited by
- ATP dependent chromatin remodeling enzymes in embryonic stem cells.
Saladi SV, de la Serna IL. Saladi SV, et al. Stem Cell Rev Rep. 2010 Mar;6(1):62-73. doi: 10.1007/s12015-010-9120-y. Stem Cell Rev Rep. 2010. PMID: 20148317 Free PMC article. Review. - An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency.
Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. Ho L, et al. Proc Natl Acad Sci U S A. 2009 Mar 31;106(13):5181-6. doi: 10.1073/pnas.0812889106. Epub 2009 Mar 11. Proc Natl Acad Sci U S A. 2009. PMID: 19279220 Free PMC article. - CHDGKB: a knowledgebase for systematic understanding of genetic variations associated with non-syndromic congenital heart disease.
Yang L, Yang Y, Liu X, Chen Y, Chen Y, Lin Y, Sun Y, Shen B. Yang L, et al. Database (Oxford). 2020 Jan 1;2020:baaa048. doi: 10.1093/database/baaa048. Database (Oxford). 2020. PMID: 32608479 Free PMC article. - Role of the Pbrm1 subunit and the PBAF complex in Schwann cell development.
Polanetzki V, Fröb F, Baroti T, Schimmel M, Tamm ER, Wegner M. Polanetzki V, et al. Sci Rep. 2022 Feb 16;12(1):2651. doi: 10.1038/s41598-022-06588-8. Sci Rep. 2022. PMID: 35173232 Free PMC article. - PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes.
Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K, Wang W. Yan Z, et al. Genes Dev. 2005 Jul 15;19(14):1662-7. doi: 10.1101/gad.1323805. Epub 2005 Jun 28. Genes Dev. 2005. PMID: 15985610 Free PMC article.
References
- Barak Y., Nelson, M.C., Ong, E.S., Jones, Y.Z., Ruiz-Lozano, P., Chien, K.R., Koder, A., and Evans, R.M. 1999. PPAR γ is required for placental, cardiac, and adipose tissue development. Mol. Cell 4: 585-595. - PubMed
- Chen T.H., Chang, T.C., Kang, J.O., Choudhary, B., Makita, T., Tran, C.M., Burch, J.B., Eid, H., and Sucov, H.M. 2002. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Dev. Biol. 250: 198-207. - PubMed
- Chi T.H., Wan, M., Zhao, K., Taniuchi, I., Chen, L., Littman, D.R., and Crabtree, G.R. 2002. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418: 195-199. - PubMed
- de The H., Vivanco-Ruiz, M.M., Tiollais, P., Stunnenberg, H., and Dejean, A. 1990. Identification of a retinoic acid responsive element in the retinoic acid receptor β gene. Nature 343: 177-180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous