Characteristic low density and shear sensitivity of cross-linked chromatin containing polycomb complexes - PubMed (original) (raw)
Characteristic low density and shear sensitivity of cross-linked chromatin containing polycomb complexes
Yuri B Schwartz et al. Mol Cell Biol. 2005 Jan.
Abstract
Chromatin cross-linking is widely used for mapping the distribution of chromosomal proteins by immunoprecipitation, but our knowledge of the physical properties of chromatin complexes remains rudimentary. Density gradients have been long used to separate fragments of cross-linked chromatin with their bound proteins from free protein or free DNA. We find that the association of DNA fragments with very-high-molecular-weight protein complexes shifts their buoyant density to values much lower then that of bulk chromatin. We show that in a CsCl gradient, Polycomb response elements, promoters of active genes, and insulator or boundary elements are found at buoyant densities similar to those of free protein and are depleted from the bulk chromatin fractions. In these regions, the low density is associated with the presence of large protein complexes and with high sensitivity to sonication. Our results suggest that separation of different chromatin regions according to their buoyant density may bias chromatin immunoprecipitation results. Density centrifugation of cross-linked chromatin may provide a simple approach to investigate the properties of large chromatin complexes in vivo.
Figures
FIG. 1.
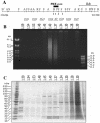
Separation of cross-linked chromatin in a CsCl density gradient. (A) Map of the bxd PRE and adjoining region of the Ubx gene. Sites for the following endonucleases are indicated: BamHI (B') AvaII (A), StyI (S), HinfI (F), EcoRI (R), BglII (B), PstI (P), KpnI (K), and HindIII (H). The coordinates of BamHI and HindIII sites according to the sequence given in reference are shown below. Note the cluster of DNase I-hypersensitive sites (arrows) in the PRE core region. The positions of the DNA fragments, whose migration in the equilibrium density gradients was analyzed by real-time PCR, are indicated below the map. Typical pictures of bulk DNA (B) and protein (C) distributions along the density gradient as assayed by gel electrophoresis. The positions of H1 and core histones are marked by black circles. The fraction densities in grams per cubic centimeter are indicated above the gels.
FIG. 2.
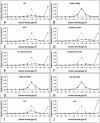
Cross-linked chromatin of the bxd PRE core has a low buoyant density. Distribution of DNA fragments from the bxd PRE region (A to H) along the same CsCl density gradient was compared to that of a DNA fragment from the coding region of the white gene (I). Here and in Fig. 3 the amount of DNA fragment in each gradient fraction (diamonds and solid line) was determined by real-time PCR using specific oligonucleotide primers and expressed as a fraction of the amount of this DNA fragment present in the entire gradient. The dotted line indicates distribution of bulk DNA estimated by gel electrophoresis and expressed in the same way. (J) Distribution of the BP fragment in a different CsCl density gradient made using very lightly cross-linked chromatin (0.3% formaldehyde).
FIG. 3.
Migration of various chromatin regions in a CsCl density gradient. Fractions of a single density gradient, different from the one used in the experiment for which results are shown in Fig. 2, were scanned by real-time PCR using oligonucleotide primers specific for different chromatin regions (diamonds and solid line). To facilitate the comparison between the two sets of experiments, the distributions of BP fragment (A) and white coding region (B) are included. The dotted line indicates the distribution of bulk DNA estimated by gel electrophoresis.
FIG. 4.
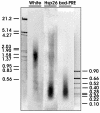
DNA fragment sizes of different regions in sonicated chromatin. The total DNA isolated from cross-linked sonicated SL2 chromatin was separated by electrophoresis, transferred to a nylon membrane, and hybridized with radioactively labeled probes from the coding region of the white gene, the bxd PRE, and the hsp26 promoter region. The positions of marker DNA fragments (sizes in kilobases) are indicated.
FIG. 5.
Cross-linked chromatin of the bxd PRE core retains its low buoyant density after intensive sonication. The chromatin cross-linked as described in the Materials and Methods was subjected to prolonged shearing (16 30-s bursts of sonication as compared to the usual 4). (A) As demonstrated by electrophoresis in a 1% agarose gel, the sizes of the DNA fragments isolated from chromatin treated in such a way (lane 2) range from 4 kbp to 100 bp, with an average size below 1 kb. Lanes 1 and 3 contain DNA size markers. (B) Real-time PCR with DNA from the fractions of the density gradient prepared from this chromatin shows that the bxd PRE (diamonds and solid line) still has a low density compared to that of the bulk chromatin or the coding region of the white gene (empty squares and dotted line).
FIG. 6.
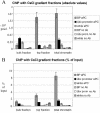
Immunoprecipitation of chromatin from different parts of the CsCl equilibrium density gradient. Two density gradients made from the same chromatin were run in parallel. Both were analyzed for the distribution of bxd PRE, white coding, and Ubx promoter regions as well as bulk DNA. The results of the analysis for one of the gradients are shown in Fig. 2. ChIP was carried out using equal volumes from the gradient fractions containing the bulk DNA peak (bulk chromatin) and the low-density material (top fraction) as well as equivalent amounts of input chromatin before ultracentrifugation (total chromatin). The absolute amounts of specific DNA fragments precipitated with anti-PC and without antibody were quantified by real-time PCR. (A) The results from the experiments with the two density gradients were averaged and plotted with standard deviations shown. (B) The same data are represented after normalization to the initial amounts of each DNA fragment in the corresponding ChIP reaction.
Similar articles
- Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation.
Nowak DE, Tian B, Brasier AR. Nowak DE, et al. Biotechniques. 2005 Nov;39(5):715-25. doi: 10.2144/000112014. Biotechniques. 2005. PMID: 16315372 - Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation.
Orlando V. Orlando V. Trends Biochem Sci. 2000 Mar;25(3):99-104. doi: 10.1016/s0968-0004(99)01535-2. Trends Biochem Sci. 2000. PMID: 10694875 Review. - Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin.
Orlando V, Paro R. Orlando V, et al. Cell. 1993 Dec 17;75(6):1187-98. doi: 10.1016/0092-8674(93)90328-n. Cell. 1993. PMID: 7903220 - Polycomb response elements and targeting of Polycomb group proteins in Drosophila.
Müller J, Kassis JA. Müller J, et al. Curr Opin Genet Dev. 2006 Oct;16(5):476-84. doi: 10.1016/j.gde.2006.08.005. Epub 2006 Aug 17. Curr Opin Genet Dev. 2006. PMID: 16914306 Review.
Cited by
- dMec: a novel Mi-2 chromatin remodelling complex involved in transcriptional repression.
Kunert N, Wagner E, Murawska M, Klinker H, Kremmer E, Brehm A. Kunert N, et al. EMBO J. 2009 Mar 4;28(5):533-44. doi: 10.1038/emboj.2009.3. Epub 2009 Jan 22. EMBO J. 2009. PMID: 19165147 Free PMC article. - Genome-wide profiling of salt fractions maps physical properties of chromatin.
Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Henikoff S, et al. Genome Res. 2009 Mar;19(3):460-9. doi: 10.1101/gr.087619.108. Epub 2008 Dec 16. Genome Res. 2009. PMID: 19088306 Free PMC article. - An evolutionary consequence of dosage compensation on Drosophila melanogaster female X-chromatin structure?
Zhang Y, Oliver B. Zhang Y, et al. BMC Genomics. 2010 Jan 5;11:6. doi: 10.1186/1471-2164-11-6. BMC Genomics. 2010. PMID: 20051121 Free PMC article. - Poly(ADP-ribosyl)ation associated changes in CTCF-chromatin binding and gene expression in breast cells.
Pavlaki I, Docquier F, Chernukhin I, Kita G, Gretton S, Clarkson CT, Teif VB, Klenova E. Pavlaki I, et al. Biochim Biophys Acta Gene Regul Mech. 2018 Aug;1861(8):718-730. doi: 10.1016/j.bbagrm.2018.06.010. Epub 2018 Jul 5. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29981477 Free PMC article. - Surveying the epigenomic landscape, one base at a time.
Zentner GE, Henikoff S. Zentner GE, et al. Genome Biol. 2012 Oct 22;13(10):250. doi: 10.1186/gb4051. Genome Biol. 2012. PMID: 23088423 Free PMC article. Review.
References
- Bollag, D. M., M. D. Rozycki, and S. J. Edelstein. 1996. Protein methods. Wiley-Liss, Inc., New York, N.Y.
- Breiling, A., B. M. Turner, M. E. Bianchi, and V. Orlando. 2001. General transcription factors bind promoters repressed by Polycomb group proteins. Nature 412:651-655. - PubMed
- Brutlag, D., C. Schlehuber, and J. Bonner. 1969. Properties of formaldehyde-treated nucleohistone. Biochemistry 8:3214-3218. - PubMed
- Cavalli, G., V. Orlando, and R. Paro. 1999. Mapping DNA target sites of chromatin-associated proteins by formaldehyde cross-linking in Drosophila embryos, p. 20-30. In W. A. Bickmore (ed.), Chromosome structural analysis: a practical approach. Oxford University Press, Oxford, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases