PDB-Ligand: a ligand database based on PDB for the automated and customized classification of ligand-binding structures - PubMed (original) (raw)
PDB-Ligand: a ligand database based on PDB for the automated and customized classification of ligand-binding structures
Jae-Min Shin et al. Nucleic Acids Res. 2005.
Abstract
PDB-Ligand (http://www.idrtech.com/PDB-Ligand/) is a three-dimensional structure database of small molecular ligands that are bound to larger biomolecules deposited in the Protein Data Bank (PDB). It is also a database tool that allows one to browse, classify, superimpose and visualize these structures. As of May 2004, there are about 4870 types of small molecular ligands, experimentally determined as a complex with protein or DNA in the PDB. The proteins that a given ligand binds are often homologous and present the same binding structure to the ligand. However, there are also many instances wherein a given ligand binds to two or more unrelated proteins, or to the same or homologous protein in different binding environments. PDB-Ligand serves as an interactive structural analysis and clustering tool for all the ligand-binding structures in the PDB. PDB-Ligand also provides an easier way to obtain a number of different structure alignments of many related ligand-binding structures based on a simple and flexible ligand clustering method. PDB-Ligand will be a good resource for both a better interpretation of ligand-binding structures and the development of better scoring functions to be used in many drug discovery applications.
Figures
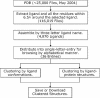
Figure 1
Scheme used in the PDB-Ligand database construction.
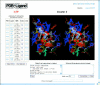
Figure 2
A typical ATP-ligand cluster, obtained using default options. The 10 ATP-binding structures from 10 PDB entries are displayed in the ligand-information table on the left-hand side. The superimposed ATP-binding structures are shown in the chime-window in stereo view. ATP is shown in CPK model and the surrounding amino acids as connected line segments in different colors depending on the residue type. The chime utilities were used for the graphics rendering.
Similar articles
- sc-PDB: an annotated database of druggable binding sites from the Protein Data Bank.
Kellenberger E, Muller P, Schalon C, Bret G, Foata N, Rognan D. Kellenberger E, et al. J Chem Inf Model. 2006 Mar-Apr;46(2):717-27. doi: 10.1021/ci050372x. J Chem Inf Model. 2006. PMID: 16563002 - Domain-based small molecule binding site annotation.
Snyder KA, Feldman HJ, Dumontier M, Salama JJ, Hogue CW. Snyder KA, et al. BMC Bioinformatics. 2006 Mar 17;7:152. doi: 10.1186/1471-2105-7-152. BMC Bioinformatics. 2006. PMID: 16545112 Free PMC article. - Identification of the ligand binding sites on the molecular surface of proteins.
Kinoshita K, Nakamura H. Kinoshita K, et al. Protein Sci. 2005 Mar;14(3):711-8. doi: 10.1110/ps.041080105. Epub 2005 Feb 2. Protein Sci. 2005. PMID: 15689509 Free PMC article. - The use of protein-ligand interaction fingerprints in docking.
Brewerton SC. Brewerton SC. Curr Opin Drug Discov Devel. 2008 May;11(3):356-64. Curr Opin Drug Discov Devel. 2008. PMID: 18428089 Review. - A structural classification of substrate-binding proteins.
Berntsson RP, Smits SH, Schmitt L, Slotboom DJ, Poolman B. Berntsson RP, et al. FEBS Lett. 2010 Jun 18;584(12):2606-17. doi: 10.1016/j.febslet.2010.04.043. Epub 2010 Apr 20. FEBS Lett. 2010. PMID: 20412802 Review.
Cited by
- Medicine for chronic atrophic gastritis: a systematic review, meta- and network pharmacology analysis.
Weng J, Wu XF, Shao P, Liu XP, Wang CX. Weng J, et al. Ann Med. 2023;55(2):2299352. doi: 10.1080/07853890.2023.2299352. Epub 2024 Jan 3. Ann Med. 2023. PMID: 38170849 Free PMC article. - Functional Evolution of Proteins.
Catazaro J, Caprez A, Swanson D, Powers R. Catazaro J, et al. Proteins. 2019 Jun;87(6):492-501. doi: 10.1002/prot.25670. Epub 2019 Feb 19. Proteins. 2019. PMID: 30714210 Free PMC article. - RPocket: an intuitive database of RNA pocket topology information with RNA-ligand data resources.
Zhou T, Wang H, Zeng C, Zhao Y. Zhou T, et al. BMC Bioinformatics. 2021 Sep 8;22(1):428. doi: 10.1186/s12859-021-04349-4. BMC Bioinformatics. 2021. PMID: 34496744 Free PMC article. - Three dimensional model of severe acute respiratory syndrome coronavirus helicase ATPase catalytic domain and molecular design of severe acute respiratory syndrome coronavirus helicase inhibitors.
Hoffmann M, Eitner K, von Grotthuss M, Rychlewski L, Banachowicz E, Grabarkiewicz T, Szkoda T, Kolinski A. Hoffmann M, et al. J Comput Aided Mol Des. 2006 May;20(5):305-19. doi: 10.1007/s10822-006-9057-z. Epub 2006 Sep 14. J Comput Aided Mol Des. 2006. PMID: 16972168 Free PMC article. - GSP4PDB: a web tool to visualize, search and explore protein-ligand structural patterns.
Angles R, Arenas-Salinas M, García R, Reyes-Suarez JA, Pohl E. Angles R, et al. BMC Bioinformatics. 2020 Mar 11;21(Suppl 2):85. doi: 10.1186/s12859-020-3352-x. BMC Bioinformatics. 2020. PMID: 32164553 Free PMC article.
References
- Holm L. and Sander,C. (1996) Mapping the protein universe. Science, 273, 595–602. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials