The UCSC Proteome Browser - PubMed (original) (raw)
The UCSC Proteome Browser
Fan Hsu et al. Nucleic Acids Res. 2005.
Abstract
The University of California Santa Cruz (UCSC) Proteome Browser provides a wealth of protein information presented in graphical images and with links to other protein-related Internet sites. The Proteome Browser is tightly integrated with the UCSC Genome Browser. For the first time, Genome Browser users have both the genome and proteome worlds at their fingertips simultaneously. The Proteome Browser displays tracks of protein and genomic sequences, exon structure, polarity, hydrophobicity, locations of cysteine and glycosylation potential, Superfamily domains and amino acids that deviate from normal abundance. Histograms show genome-wide distribution of protein properties, including isoelectric point, molecular weight, number of exons, InterPro domains and cysteine locations, together with specific property values of the selected protein. The Proteome Browser also provides links to gene annotations in the Genome Browser, the Known Genes details page and the Gene Sorter; domain information from Superfamily, InterPro and Pfam; three-dimensional structures at the Protein Data Bank and ModBase; and pathway data at KEGG, BioCarta/CGAP and BioCyc. As of August 2004, the Proteome Browser is available for human, mouse and rat proteomes. The browser may be accessed from any Known Genes details page of the Genome Browser at http://genome.ucsc.edu. A user's guide is also available on this website.
Figures
Figure 1
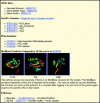
This Proteome Browser tracks display sample shows the tracks of human protein CLOC_HUMAN in DNA mode that display both the protein and genomic sequences. As the corresponding mRNA is a negative strand, the DNA sequence and its complementary coding sequence are both shown. On the exon track, a part of the 2nd and 3rd exons is shown. The start of a Superfamily domain, Helix–loop–helix DNA-binding domain, is shown by the yellow bar. In this particular protein, the amino acid glutamine (Q) is flagged to indicate that it is present in a higher-than-average percentage.
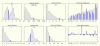
Figure 2
The Proteome Browser property histograms sample shows that this protein has high molecular weight and a large number of exons. It has four InterPro domains and an unusually high concentration of glutamine.
Figure 3
This example shows the web links to UCSC Genome Browser, Known Gene details page and Gene Sorter; InterPro and Pfam domains; predicted 3D structure by ModBase; and pathways from BioCarta and KEGG.
Similar articles
- The UCSC Genome Browser Database: update 2006.
Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, Hillman-Jackson J, Kuhn RM, Pedersen JS, Pohl A, Raney BJ, Rosenbloom KR, Siepel A, Smith KE, Sugnet CW, Sultan-Qurraie A, Thomas DJ, Trumbower H, Weber RJ, Weirauch M, Zweig AS, Haussler D, Kent WJ. Hinrichs AS, et al. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D590-8. doi: 10.1093/nar/gkj144. Nucleic Acids Res. 2006. PMID: 16381938 Free PMC article. - The UCSC Known Genes.
Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. Hsu F, et al. Bioinformatics. 2006 May 1;22(9):1036-46. doi: 10.1093/bioinformatics/btl048. Epub 2006 Feb 24. Bioinformatics. 2006. PMID: 16500937 - The UCSC Table Browser data retrieval tool.
Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. Karolchik D, et al. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D493-6. doi: 10.1093/nar/gkh103. Nucleic Acids Res. 2004. PMID: 14681465 Free PMC article. - UCSC genome browser tutorial.
Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ. Zweig AS, et al. Genomics. 2008 Aug;92(2):75-84. doi: 10.1016/j.ygeno.2008.02.003. Epub 2008 Jun 2. Genomics. 2008. PMID: 18514479 Review. - The genome browser at UCSC for locating genes, and much more!
Bina M. Bina M. Mol Biotechnol. 2008 Mar;38(3):269-75. doi: 10.1007/s12033-007-9019-2. Epub 2007 Dec 4. Mol Biotechnol. 2008. PMID: 18058261 Review.
Cited by
- NCATS Inxight Drugs: a comprehensive and curated portal for translational research.
Siramshetty VB, Grishagin I, Nguyễn ÐT, Peryea T, Skovpen Y, Stroganov O, Katzel D, Sheils T, Jadhav A, Mathé EA, Southall NT. Siramshetty VB, et al. Nucleic Acids Res. 2022 Jan 7;50(D1):D1307-D1316. doi: 10.1093/nar/gkab918. Nucleic Acids Res. 2022. PMID: 34648031 Free PMC article. - The UCSC genome browser database: update 2007.
Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pedersen JS, Hsu F, Hinrichs AS, Harte RA, Diekhans M, Clawson H, Bejerano G, Barber GP, Baertsch R, Haussler D, Kent WJ. Kuhn RM, et al. Nucleic Acids Res. 2007 Jan;35(Database issue):D668-73. doi: 10.1093/nar/gkl928. Epub 2006 Nov 16. Nucleic Acids Res. 2007. PMID: 17142222 Free PMC article. - Genomic location of the human RNA polymerase II general machinery: evidence for a role of TFIIF and Rpb7 at both early and late stages of transcription.
Cojocaru M, Jeronimo C, Forget D, Bouchard A, Bergeron D, Côte P, Poirier GG, Greenblatt J, Coulombe B. Cojocaru M, et al. Biochem J. 2008 Jan 1;409(1):139-47. doi: 10.1042/BJ20070751. Biochem J. 2008. PMID: 17848138 Free PMC article. - Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes.
Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, Ishii S, Sugiyama T, Saito K, Isono Y, Irie R, Kushida N, Yoneyama T, Otsuka R, Kanda K, Yokoi T, Kondo H, Wagatsuma M, Murakawa K, Ishida S, Ishibashi T, Takahashi-Fujii A, Tanase T, Nagai K, Kikuchi H, Nakai K, Isogai T, Sugano S. Kimura K, et al. Genome Res. 2006 Jan;16(1):55-65. doi: 10.1101/gr.4039406. Epub 2005 Dec 12. Genome Res. 2006. PMID: 16344560 Free PMC article. - ProteomeScout: a repository and analysis resource for post-translational modifications and proteins.
Matlock MK, Holehouse AS, Naegle KM. Matlock MK, et al. Nucleic Acids Res. 2015 Jan;43(Database issue):D521-30. doi: 10.1093/nar/gku1154. Epub 2014 Nov 20. Nucleic Acids Res. 2015. PMID: 25414335 Free PMC article.
References
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials