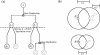Inparanoid: a comprehensive database of eukaryotic orthologs - PubMed (original) (raw)
Inparanoid: a comprehensive database of eukaryotic orthologs
Kevin P O'Brien et al. Nucleic Acids Res. 2005.
Abstract
The Inparanoid eukaryotic ortholog database (http://inparanoid.cgb.ki.se/) is a collection of pairwise ortholog groups between 17 whole genomes; Anopheles gambiae, Caenorhabditis briggsae, Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, Takifugu rubripes, Gallus gallus, Homo sapiens, Mus musculus, Pan troglodytes, Rattus norvegicus, Oryza sativa, Plasmodium falciparum, Arabidopsis thaliana, Escherichia coli, Saccharomyces cerevisiae and Schizosaccharomyces pombe. Complete proteomes for these genomes were derived from Ensembl and UniProt and compared pairwise using Blast, followed by a clustering step using the Inparanoid program. An Inparanoid cluster is seeded by a reciprocally best-matching ortholog pair, around which inparalogs (should they exist) are gathered independently, while outparalogs are excluded. The ortholog clusters can be searched on the website using Ensembl gene/protein or UniProt identifiers, annotation text or by Blast alignment against our protein datasets. The entire dataset can be downloaded, as can the Inparanoid program itself.
Figures
Figure 1
A hypothetical gene tree and the resulting Inparanoid clusters are shown to illustrate inparalog (and thus co-ortholog) and outparalog assignments. (a) Protein A in an ancestral species ‘A’ undergoes a gene duplication. A speciation event occurs which gives rise to the two lineages leading to species ‘B’ and ‘C’. In the C genome the genes C2 and C3 are inparalogs since their gene duplication occurred after speciation; they are co-orthologous to the B2 gene (one common ancestral protein upon speciation). B1 is an outparalog of the C2 and C3 genes, as are B1 of B2 (duplication and divergence prior to speciation). (b) B2 and C2 are the original seed-ortholog pair (all inparalogs are clustered around this pair), thus both receiving an inparalog score of 1.0. Other inparalogs (in this case C3) are scored according to their relative similarity to the seed-inparalog (here C2). Inparalog score of C3 = (Blast[C2:_C3_]−Blast[C2:_B2_])/(Blast[C2:_C2_]−Blast[C2:_B2_]) where Blast[X:Y] is the averaged blast score between X and Y in bits. In this case C2 is relatively more similar to B2 than C3 is, and thus C3 receives a lower inparalog score (0.7). C1 and B1 are orthologous to each other but are outparalogs of the other cluster and thus form a cluster of their own.
Figure 2
An Inparanoid cluster is a representation of genes thought to share a single ancestral gene upon speciation. In this example output only human–mosquito and human–worm clusters are shown. In the human–worm cluster, the two human genes are inparalogs, i.e. resulted from a gene duplication after the speciation from worm occurred. They are thus both co-orthologous to the worm gene. The Inparanoid score is a measure of how similar an inparalog is to the inparalog that is the main ortholog. If they are identical the score is 1.0, but as the similarity drops towards the similarity of the main orthologs, the score goes to 0.0 (see Figure 1). For example, ENSP00000343386 is less similar to the C.briggsae gene than is ENSP00000322439, but both these human genes are still co-orthologous to it. The bootstrapping score is a measure of how reliably that gene is the main ortholog. Gene and protein identifiers are hyperlinks to the relevant databases for each species.
Figure 3
Using a gene name as a search generates a list of possible gene hits. Clicking on the ‘Search for Clusters’ icon queries the database as to whether a gene occurs in an Inparanoid-cluster in all organisms. Clicking on the cluster name generates a multiple FASTA file for this cluster and performs a multiple alignment using the Kalign program. These can be used to check the validity of the cluster in question and can be saved to disk.
Similar articles
- OrthoDisease: a database of human disease orthologs.
O'Brien KP, Westerlund I, Sonnhammer EL. O'Brien KP, et al. Hum Mutat. 2004 Aug;24(2):112-9. doi: 10.1002/humu.20068. Hum Mutat. 2004. PMID: 15241792 - Automatic clustering of orthologs and inparalogs shared by multiple proteomes.
Alexeyenko A, Tamas I, Liu G, Sonnhammer EL. Alexeyenko A, et al. Bioinformatics. 2006 Jul 15;22(14):e9-15. doi: 10.1093/bioinformatics/btl213. Bioinformatics. 2006. PMID: 16873526 - eSLDB: eukaryotic subcellular localization database.
Pierleoni A, Martelli PL, Fariselli P, Casadio R. Pierleoni A, et al. Nucleic Acids Res. 2007 Jan;35(Database issue):D208-12. doi: 10.1093/nar/gkl775. Epub 2006 Nov 15. Nucleic Acids Res. 2007. PMID: 17108361 Free PMC article. - Automatic clustering of orthologs and in-paralogs from pairwise species comparisons.
Remm M, Storm CE, Sonnhammer EL. Remm M, et al. J Mol Biol. 2001 Dec 14;314(5):1041-52. doi: 10.1006/jmbi.2000.5197. J Mol Biol. 2001. PMID: 11743721 - Orthology and functional conservation in eukaryotes.
Dolinski K, Botstein D. Dolinski K, et al. Annu Rev Genet. 2007;41:465-507. doi: 10.1146/annurev.genet.40.110405.090439. Annu Rev Genet. 2007. PMID: 17678444 Review.
Cited by
- Multi-omic analysis of bat versus human fibroblasts reveals altered central metabolism.
Jagannathan NS, Koh JYP, Lee Y, Sobota RM, Irving AT, Wang LF, Itahana Y, Itahana K, Tucker-Kellogg L. Jagannathan NS, et al. Elife. 2024 Jul 22;13:e94007. doi: 10.7554/eLife.94007. Elife. 2024. PMID: 39037770 Free PMC article. - ToxoNet: A high confidence map of protein-protein interactions in Toxoplasma gondii.
Swapna LS, Stevens GC, Sardinha-Silva A, Hu LZ, Brand V, Fusca DD, Wan C, Xiong X, Boyle JP, Grigg ME, Emili A, Parkinson J. Swapna LS, et al. PLoS Comput Biol. 2024 Jun 20;20(6):e1012208. doi: 10.1371/journal.pcbi.1012208. eCollection 2024 Jun. PLoS Comput Biol. 2024. PMID: 38900844 Free PMC article. - Improved integration of single-cell transcriptome data demonstrates common and unique signatures of heart failure in mice and humans.
Jurado MR, Tombor LS, Arsalan M, Holubec T, Emrich F, Walther T, Abplanalp W, Fischer A, Zeiher AM, Schulz MH, Dimmeler S, John D. Jurado MR, et al. Gigascience. 2024 Jan 2;13:giae011. doi: 10.1093/gigascience/giae011. Gigascience. 2024. PMID: 38573186 Free PMC article. - Seagrass genomes reveal ancient polyploidy and adaptations to the marine environment.
Ma X, Vanneste S, Chang J, Ambrosino L, Barry K, Bayer T, Bobrov AA, Boston L, Campbell JE, Chen H, Chiusano ML, Dattolo E, Grimwood J, He G, Jenkins J, Khachaturyan M, Marín-Guirao L, Mesterházy A, Muhd DD, Pazzaglia J, Plott C, Rajasekar S, Rombauts S, Ruocco M, Scott A, Tan MP, Van de Velde J, Vanholme B, Webber J, Wong LL, Yan M, Sung YY, Novikova P, Schmutz J, Reusch TBH, Procaccini G, Olsen JL, Van de Peer Y. Ma X, et al. Nat Plants. 2024 Feb;10(2):240-255. doi: 10.1038/s41477-023-01608-5. Epub 2024 Jan 26. Nat Plants. 2024. PMID: 38278954 Free PMC article. - Resistance gene-guided genome mining reveals the roseopurpurins as inhibitors of cyclin-dependent kinases.
Dunbar KL, Perlatti B, Liu N, Cornelius A, Mummau D, Chiang YM, Hon L, Nimavat M, Pallas J, Kordes S, Ng HL, Harvey CJB. Dunbar KL, et al. Proc Natl Acad Sci U S A. 2023 Nov 28;120(48):e2310522120. doi: 10.1073/pnas.2310522120. Epub 2023 Nov 20. Proc Natl Acad Sci U S A. 2023. PMID: 37983497 Free PMC article.
References
- Remm M., Storm,C.E. and Sonnhammer,E.L. (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol., 314, 1041–1052. - PubMed
- O'Brien K.P., Westerlund,I. and Sonnhammer,E.L. (2004) OrthoDisease: a database of human disease orthologs. Hum. Mutat., 24, 112–119. - PubMed
- Sonnhammer E.L. and Koonin,E.V. (2002) Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet., 18, 619–620. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous