A complex zinc finger controls the enzymatic activities of nidovirus helicases - PubMed (original) (raw)
A complex zinc finger controls the enzymatic activities of nidovirus helicases
Anja Seybert et al. J Virol. 2005 Jan.
Abstract
Nidoviruses (Coronaviridae, Arteriviridae, and Roniviridae) encode a nonstructural protein, called nsp10 in arteriviruses and nsp13 in coronaviruses, that is comprised of a C-terminal superfamily 1 helicase domain and an N-terminal, putative zinc-binding domain (ZBD). Previously, mutations in the equine arteritis virus (EAV) nsp10 ZBD were shown to block arterivirus reproduction by disrupting RNA synthesis and possibly virion biogenesis. Here, we characterized the ATPase and helicase activities of bacterially expressed mutant forms of nsp10 and its human coronavirus 229E ortholog, nsp13, and correlated these in vitro activities with specific virus phenotypes. Replacement of conserved Cys or His residues with Ala proved to be more deleterious than Cys-for-His or His-for-Cys replacements. Furthermore, denaturation-renaturation experiments revealed that, during protein refolding, Zn2+ is essential for the rescue of the enzymatic activities of nidovirus helicases. Taken together, the data strongly support the zinc-binding function of the N-terminal domain of nidovirus helicases. nsp10 ATPase/helicase deficiency resulting from single-residue substitutions in the ZBD or deletion of the entire domain could not be complemented in trans by wild-type ZBD, suggesting a critical function of the ZBD in cis. Consistently, no viral RNA synthesis was detected after transfection of EAV full-length RNAs encoding ATPase/helicase-deficient nsp10 into susceptible cells. In contrast, diverse phenotypes were observed for mutants with enzymatically active nsp10, which in a number of cases correlated with the activities measured in vitro. Collectively, our data suggest that the ZBD is critically involved in nidovirus replication and transcription by modulating the enzymatic activities of the helicase domain and other, yet unknown, mechanisms.
Figures
FIG. 1.
Sequences of the N-terminal ZBDs associated with the arterivirus EAV nsp10 and coronavirus HCoV-229E nsp13 helicases. Amino acid residues are numbered according to their positions in the pp1ab replicase polyproteins of EAV and HCoV-229E, respectively. 3C-like protease cleavage sites that flank the EAV and HCoV-229E helicases in pp1ab are given, and the hinge spacer region of nsp10 (residues Glu2427 to Pro2430) is underlined. Also indicated are the amino acid substitutions characterized in this study.
FIG. 2.
Effects of substitutions of conserved ZBD Cys and His residues on the enzymatic activities of EAV nsp10 in vitro and on EAV reproduction in BHK-21 cells. Helicase (duplex-unwinding) activities were determined by using a forked DNA substrate containing a 22-bp duplex region and 30-nucleotide, single-stranded oligo(dT) tails (24). This partial-duplex DNA substrate was incubated with 2 pmol (if not indicated otherwise) of purified MBP-nsp10 or its mutant derivatives (for details, see Materials and Methods). Reaction products were analyzed on nondenaturing polyacrylamide gels. Lane 1, incubation without protein; lane 2, heat-denatured substrate; lane 3, MBP-nsp10 (wild type [WT]); lane 4, MBP-nsp10_C2377H; lane 5, MBP-nsp10_C2395H; lane 6, MBP-nsp10_C2395A; lane 7, MBP-nsp10_H2399C; lane 8, MBP-nsp10_H2399A; lane 9, MBP-nsp10_C2412H; lane 10, MBP-nsp10_H2414C; lane 11, MBP-nsp10_H2414A; lane 12, MBP-nsp10_K2534Q; lane 13, 1 pmol of MBP-nsp10_K2534Q and 1 pmol of MBP-nsp10_C2377H; lane 14, MBP-nsp10_K2534Q and MBP-nsp10_C2377H; lane 15, 1 pmol of MBP-nsp10_K2534Q and 1 pmol of MBP-nsp10_H2399C. Below the gel, the relative ATPase activities of the corresponding proteins are given. The activity of the wild-type protein, MBP-nsp10, was taken to be 1.0, and all other ATPase activities were normalized to this value. Also listed are the phenotypes in tissue culture of EAV mutants containing the respective nsp10 ZBD substitutions (Table 1) (35). Titers of progeny virus were determined from tissue culture supernatants harvested at 24 h posttransfection. RNA replication and sgRNA synthesis, respectively, were assessed by immunofluorescence analysis by using antibodies specific for the replicase gene product, nsp3, and the nucleocapsid protein, respectively (for details, see Materials and Methods). RNA synthesis of the EAV mutants, EAV_C2395H and EAV_H2414C, which had delayed growth kinetics, was also studied by Northern blotting (Fig. 3). −, absent; +, present; dsDNA, double-stranded DNA; ssDNA, single-stranded DNA.
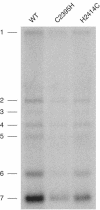
FIG. 3.
Hybridization analysis of the RNA synthesis of EAV nsp10 mutants EAV_C2395H and EAV_H2414C. Intracellular RNA was isolated from similar numbers of infected cells at 12 h postinfection. Both genome replication and sgRNA synthesis of the two mutants were reduced compared to a wild-type (WT) control. EAV RNAs 1 to 7 are indicated to the left.
FIG. 4.
Effects of substitutions and deletions in the EAV nsp10 hinge spacer region. Helicase activity assays were carried out under the same conditions as in the experiment shown in Fig. 2. Lane 1, reaction without protein; lane 2, heat-denatured substrate; lane 3, MBP-nsp10 (wild type [WT]); lane 4, MBP-nsp10_K2534Q; lane 5, MBP-nsp10_S2429P; lane 6, MBP-nsp10_S2429G/P2430G; lane 7, MBP-nsp10_S2429P/P2430S; lane 8, MBP-nsp10_ΔE2427; lane 9, MBP-nsp10_ΔG2428. Below the gel, the relative ATPase activities of the respective proteins are given. The ATPase activity of the wild-type protein, MBP-nsp10, was taken to be 1.0, and all other activities were normalized to this value. In the bottom panel, the tissue culture phenotypes are given for the EAV mutants containing the respective substitutions or deletions in the hinge spacer region (35). −, absent; +, present; ±, strongly reduced; dsDNA, double-stranded DNA; ssDNA, single-stranded DNA.
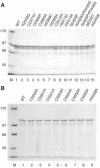
FIG. 5.
Purification of bacterially expressed MBP-EAV nsp10 and MBP-HCoV-229E nsp13 fusion proteins. Fusion proteins (1 μg each) were separated by SDS-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. Lane M shows the molecular mass markers. (A) MBP-nsp10 fusion proteins. Lane 1, wild-type (WT) MBP-nsp10; lanes 2 to 15, mutant derivatives of MBP-nsp10, with the respective amino acid substitutions indicated above the gel. (B) MBP-nsp13 fusion proteins. Lane 1, MBP-nsp13; lanes 2 to 9, mutant derivatives of MBP-nsp13, with the respective amino acid substitutions indicated above the gel.
FIG. 6.
Zn2+ is an essential structural cofactor for the ATPase activities of arterivirus EAV nsp10 and coronavirus HCoV-229E nsp13. MBP-nsp10 and MBP-nsp13 were denatured in urea-containing buffer and subsequently renatured in buffer containing zinc acetate and EDTA, respectively. The ATPase activities of the renatured MBP-nsp10 (lanes 2 and 3) and MBP-nsp13 (lanes 5 and 6) fusion proteins were analyzed by the [γ-32P]ATP thin-layer chromatography assay described in Materials and Methods. ATPase reactions were done with buffer alone (lanes 1 and 4), with proteins renatured in the presence of 100 μM zinc acetate (ZnOAc, lanes 2 and 5), and with proteins renatured in the presence of 10 mM EDTA (lanes 3 and 6). The positions of ATP and Pi are indicated.
Similar articles
- Structural Characterization of the Helicase nsp10 Encoded by Porcine Reproductive and Respiratory Syndrome Virus.
Shi Y, Tong X, Ye G, Xiu R, Li L, Sun L, Shi J, Li M, Song Y, Fan C, Shi K, Fu ZF, Xiao S, Peng G. Shi Y, et al. J Virol. 2020 Jul 16;94(15):e02158-19. doi: 10.1128/JVI.02158-19. Print 2020 Jul 16. J Virol. 2020. PMID: 32461315 Free PMC article. - Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Deng Z, Lehmann KC, Li X, Feng C, Wang G, Zhang Q, Qi X, Yu L, Zhang X, Feng W, Wu W, Gong P, Tao Y, Posthuma CC, Snijder EJ, Gorbalenya AE, Chen Z. Deng Z, et al. Nucleic Acids Res. 2014 Mar;42(5):3464-77. doi: 10.1093/nar/gkt1310. Epub 2013 Dec 24. Nucleic Acids Res. 2014. PMID: 24369429 Free PMC article. - Biochemical characterization of the equine arteritis virus helicase suggests a close functional relationship between arterivirus and coronavirus helicases.
Seybert A, van Dinten LC, Snijder EJ, Ziebuhr J. Seybert A, et al. J Virol. 2000 Oct;74(20):9586-93. doi: 10.1128/jvi.74.20.9586-9593.2000. J Virol. 2000. PMID: 11000230 Free PMC article. - What we know but do not understand about nidovirus helicases.
Lehmann KC, Snijder EJ, Posthuma CC, Gorbalenya AE. Lehmann KC, et al. Virus Res. 2015 Apr 16;202:12-32. doi: 10.1016/j.virusres.2014.12.001. Epub 2014 Dec 8. Virus Res. 2015. PMID: 25497126 Free PMC article. Review. - Nidovirus RNA polymerases: Complex enzymes handling exceptional RNA genomes.
Posthuma CC, Te Velthuis AJW, Snijder EJ. Posthuma CC, et al. Virus Res. 2017 Apr 15;234:58-73. doi: 10.1016/j.virusres.2017.01.023. Epub 2017 Feb 6. Virus Res. 2017. PMID: 28174054 Free PMC article. Review.
Cited by
- Host and Viral Zinc-Finger Proteins in COVID-19.
Esposito S, D'Abrosca G, Antolak A, Pedone PV, Isernia C, Malgieri G. Esposito S, et al. Int J Mol Sci. 2022 Mar 28;23(7):3711. doi: 10.3390/ijms23073711. Int J Mol Sci. 2022. PMID: 35409070 Free PMC article. Review. - An arginine-to-proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells.
Fang S, Chen B, Tay FP, Ng BS, Liu DX. Fang S, et al. Virology. 2007 Feb 5;358(1):136-47. doi: 10.1016/j.virol.2006.08.020. Epub 2006 Sep 18. Virology. 2007. PMID: 16979681 Free PMC article. - A nidovirus perspective on SARS-CoV-2.
Gulyaeva AA, Gorbalenya AE. Gulyaeva AA, et al. Biochem Biophys Res Commun. 2021 Jan 29;538:24-34. doi: 10.1016/j.bbrc.2020.11.015. Epub 2020 Nov 13. Biochem Biophys Res Commun. 2021. PMID: 33413979 Free PMC article. Review. - Synergistic Inhibition of SARS-CoV-2 Replication Using Disulfiram/Ebselen and Remdesivir.
Chen T, Fei CY, Chen YP, Sargsyan K, Chang CP, Yuan HS, Lim C. Chen T, et al. ACS Pharmacol Transl Sci. 2021 Mar 26;4(2):898-907. doi: 10.1021/acsptsci.1c00022. eCollection 2021 Apr 9. ACS Pharmacol Transl Sci. 2021. PMID: 33855277 Free PMC article. - Equine arteritis virus.
Balasuriya UB, Go YY, MacLachlan NJ. Balasuriya UB, et al. Vet Microbiol. 2013 Nov 29;167(1-2):93-122. doi: 10.1016/j.vetmic.2013.06.015. Epub 2013 Jul 3. Vet Microbiol. 2013. PMID: 23891306 Free PMC article. Review.
References
- Aubry, F., M. G. Mattei, and F. Galibert. 1998. Identification of a human 17p-located cDNA encoding a protein of the Snf2-like helicase family. Eur. J. Biochem. 254:558-564. - PubMed
- Biswas, N., and S. K. Weller. 1999. A mutation in the C-terminal putative Zn2+ finger motif of UL52 severely affects the biochemical activities of the HSV-1 helicase-primase subcomplex. J. Biol. Chem. 274:8068-8076. - PubMed
- Davidson, A., and S. Siddell. 2003. Potential for antiviral treatment of severe acute respiratory syndrome. Curr. Opin. Infect. Dis. 16:565-571. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources