Integrating reptilian herpesviruses into the family herpesviridae - PubMed (original) (raw)
Integrating reptilian herpesviruses into the family herpesviridae
Duncan J McGeoch et al. J Virol. 2005 Jan.
Abstract
The phylogeny of reptilian herpesviruses (HVs) relative to mammalian and avian HVs was investigated by using available gene sequences and by alignment of encoded amino acid sequences and derivation of trees by maximum-likelihood and Bayesian methods. Phylogenetic loci were obtained for green turtle HV (GTHV) primarily on the basis of DNA polymerase (POL) and DNA binding protein sequences, and for lung-eye-trachea disease-associated HV (LETV) primarily from its glycoprotein B sequence; both have nodes on the branch leading to recognized species in the Alphaherpesvirinae subfamily and should be regarded as new members of that subfamily. A similar but less well defined locus was obtained for an iguanid HV based on a partial POL sequence. On the basis of short POL sequences (around 60 amino acid residues), it appeared likely that GTHV and LETV belong to a private clade and that three HVs of gerrhosaurs (plated lizards) are associated with the iguanid HV. Based on phylogenetic branching patterns for mammalian HV lineages that mirror those of host lineages, we estimated a date for the HV tree's root of around 400 million years ago. Estimated dates for branching events in the development of reptilian, avian, and mammalian Alphaherpesvirinae lineages could plausibly be accounted for in part but not completely by ancient coevolution of these virus lines with reptilian lineages and with the development of birds and mammals from reptilian progenitors.
Figures
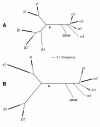
FIG. 1.
POL and DBP trees, including GTHV. The top-scoring trees are shown based on HV POL amino acid sequences (A) and HV DBP amino acid sequences (B). The trees are presented as unrooted and in a condensed format showing genus-level groupings rather than individual species, with the multiple-branch region of each genus-level clade represented by a single heavy line. In each tree, the estimated position of the root is indicated by a filled arrowhead, calculated as the midpoint of the distance from the mean positions of branch tips in the alpha subfamily to the mean positions of branch tips in the beta and gamma subfamilies. A common scale bar is indicated for divergence (i.e., substitutions per amino acid site). Genus-equivalent labels are as follows: α1, Simplexvirus; α2, Varicellovirus; α3, Mardivirus; α4, Iltovirus; β1, Cytomegalovirus (including Muromegalovirus and Tupaia HV); β2, Roseolovirus (including porcine cytomegalovirus); β3, elephant endothelial HV; γ1, Lymphocryptovirus; γ2, Rhadinovirus. (A) The POL tree contains 44 species in addition to GTHV, distributed as follows: α1, 4 species; α2, 7 species; α3, 3 species; α4, 1 species; β1, 8 species; β2, 3 species; β3, 1 species; γ1, 3 species; γ2, 14 species. (B) The DBP tree contains 35 species in addition to GTHV, distributed as follows: α1, 4 species; α2, 6 species; α3, 3 species; α4, 1 species; β1, 7 species; β2, 2 species; γ1, 3 species; γ2, 9 species.
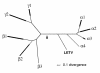
FIG. 2.
gB tree, including LETV. The top-scoring tree is shown based on HV gB amino acid sequences, obtained by two separate methods (Codeml and MrBayes). The format is as described for Fig. 1. The gB tree contains 60 species in addition to LETV, distributed as follows: α1, 10 species; α2, 13 species; α3, 3 species; α4, 1 species; β1, 9 species; β2, 3 species; β3, 1 species; γ1, 6 species; γ2, 14 species.
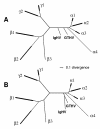
FIG. 3.
Trees based on a part of POL, including IgHV. The two top equal trees are shown based on a 226-amino-acid alignment of a section of POL. The alignment included 46 species: 45 as described for Fig. 1A plus IgHV. The format is as described for Fig. 1.
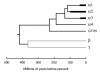
FIG. 4.
Molecular clock tree based on DBP plus POL sequences. A concatenated alignment for DBP and POL amino acid sequences of 36 species was used to produce a molecular clock, and a timescale was applied based on the correlation of divergence in the α2 lineage with paleontological dates as described in the text. Regions in the α1, α2, and α3 lineages occupied by multiple branches are shown as heavy lines, and the reduced outlines for the beta and gamma lineages are shown in gray.
FIG. 5.
Comparison of host and alphaherpesvirus trees. (A) Consensus tree for reptilian, avian, and mammalian lineages, derived from published data, with branch names chosen for comparison with the HV tree. The gray bar represents multiple branchings treated as unresolved. (B) Summary tree for alphaherpesvirus lineages, based on the gB tree and DBP plus POL molecular clock tree (Table 2). The gray bar and branch represent unresolved branching details for GTHV, LETV, and IgHV. In both trees, nodes that are discussed in the text are labeled. A common timescale is shown at the foot. inc., includes.
Similar articles
- Nucleotide sequence of an ICP18.5 assembly protein (UL28) gene of green turtle herpesvirus pathogenically associated with green turtle fibropapilloma.
Nigro O, Alonso Aguirre A, Lu Y. Nigro O, et al. J Virol Methods. 2004 Sep 1;120(1):107-12. doi: 10.1016/j.jviromet.2004.04.011. J Virol Methods. 2004. PMID: 15234815 - Rapid acquisition of entire DNA polymerase gene of a novel herpesvirus from green turtle fibropapilloma by a genomic walking technique.
Yu Q, Hu N, Lu Y, Nerurkar VR, Yanagihara R. Yu Q, et al. J Virol Methods. 2001 Feb;91(2):183-95. doi: 10.1016/s0166-0934(00)00267-6. J Virol Methods. 2001. PMID: 11164500 - Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses.
McGeoch DJ, Cook S, Dolan A, Jamieson FE, Telford EA. McGeoch DJ, et al. J Mol Biol. 1995 Mar 31;247(3):443-58. doi: 10.1006/jmbi.1995.0152. J Mol Biol. 1995. PMID: 7714900 - Inclusion Body Herpesvirus Hepatitis in Captive Falcons in the Middle East: A Review of Clinical and Pathologic Findings.
Raghav R, Samour J. Raghav R, et al. J Avian Med Surg. 2019 Mar 1;33(1):1-6. doi: 10.1647/2018-341. J Avian Med Surg. 2019. PMID: 31124605 Review. - The evolution of protein domain repertoires: Shedding light on the origins of the Herpesviridae family.
Brito AF, Pinney JW. Brito AF, et al. Virus Evol. 2020 Feb 5;6(1):veaa001. doi: 10.1093/ve/veaa001. eCollection 2020 Jan. Virus Evol. 2020. PMID: 32042448 Free PMC article. Review.
Cited by
- A novel herpesvirus in a white stork associated with splenic and hepatic necrosis.
Mack ZE, Bonar CJ, Garner MM, Connolly MJ, Childress AL, Wellehan JFX Jr. Mack ZE, et al. J Vet Diagn Invest. 2020 May;32(3):471-475. doi: 10.1177/1040638720915539. Epub 2020 Apr 10. J Vet Diagn Invest. 2020. PMID: 32274981 Free PMC article. - Detection of classical swine fever virus E2 gene in cattle serum samples from cattle herds of Meghalaya.
Chakraborty AK, Karam A, Mukherjee P, Barkalita L, Borah P, Das S, Sanjukta R, Puro K, Ghatak S, Shakuntala I, Sharma I, Laha RG, Sen A. Chakraborty AK, et al. Virusdisease. 2018 Mar;29(1):89-95. doi: 10.1007/s13337-018-0433-9. Epub 2018 Feb 17. Virusdisease. 2018. PMID: 29607364 Free PMC article. - The order Herpesvirales.
Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. Davison AJ, et al. Arch Virol. 2009;154(1):171-7. doi: 10.1007/s00705-008-0278-4. Epub 2008 Dec 9. Arch Virol. 2009. PMID: 19066710 Free PMC article. - An MHC class I immune evasion gene of Marek׳s disease virus.
Hearn C, Preeyanon L, Hunt HD, York IA. Hearn C, et al. Virology. 2015 Jan 15;475:88-95. doi: 10.1016/j.virol.2014.11.008. Epub 2014 Nov 27. Virology. 2015. PMID: 25462349 Free PMC article. - Potential of Viruses as Environmental Etiological Factors for Non-Syndromic Orofacial Clefts.
Messias TS, Silva KCP, Silva TC, Soares S. Messias TS, et al. Viruses. 2024 Mar 27;16(4):511. doi: 10.3390/v16040511. Viruses. 2024. PMID: 38675854 Free PMC article.
References
- Adachi, J., and M. Hasegawa. 1994. The MOLPHY 2.2 package. Institute of Statistical Mathematics, Tokyo, Japan.
- Benton, M. J. 1997. Vertebrate palaeontology. Chapman & Hall, London, United Kingdom.
- Cao, Y., M. D. Sorenson, Y. Kumasawa, D. P. Mindell, and M. Hasegawa. 2000. Phylogenetic position of turtles among amniotes: evidence from mitochondrial and nuclear genes. Gene 259:139-148. - PubMed
- Davison, A. J. 2002. Evolution of the herpesviruses. Vet. Microbiol. 86:69-88. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources