Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches - PubMed (original) (raw)
Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches
Leonid Gitlin et al. J Virol. 2005 Jan.
Abstract
Short interfering RNAs (siRNAs) directed against poliovirus and other viruses effectively inhibit viral replication. Although RNA interference (RNAi) may provide the basis for specific antiviral therapies, the limitations of RNAi antiviral strategies are ill defined. Here, we show that poliovirus readily escapes highly effective siRNAs through unique point mutations within the targeted regions. Competitive analysis of the escape mutants provides insights into the basis of siRNA recognition. The RNAi machinery can tolerate mismatches but is exquisitely sensitive to mutations within the central region and the 3' end of the target sequence. Indeed, specific mutations in the target sequence resulting in G:U mismatches are sufficient for the virus to escape siRNA inhibition. However, using a pool of siRNAs to simultaneously target multiple sites in the viral genome prevents the emergence of resistant viruses. Our study uncovers the elegant precision of target recognition by the RNAi machinery and provides the basis for the development of effective RNAi-based therapies that prevent viral escape.
Figures
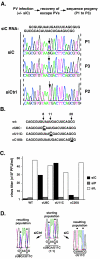
FIG. 1.
Polioviruses resistant to siC. (A) Population sequencing of the viral siC region in siC-transfected cells. siCtrl, virus grown in cells transfected with control (rhinovirus-specific) siRNA after two passages; P1, a representative viral population after one passage in siC-transfected cells; P3, P1 following two additional passages under siC. Arrows point to the newly arising mutation. (B) Mutants in the siC region, engineered into the infectious clone of poliovirus. Numbers and arrows indicate the position of each mutation. (C) Titers of wild-type and mutant viruses following infection in HeLa cells transfected with siC (leftmost black bars), siP (middle gray bars), and siL (luciferase) (white bars). Infections were done at a multiplicity of infection (MOI) of 10 and produced virus collected at 8 h postinfection. (D) Competition assay between cU11C and cU8C. cU11C and cU8C were mixed at a 1:1 ratio and passaged twice with control (siCtrl)- or siC-transfected cells. The resulting population was analyzed by sequencing an RT-PCR product corresponding to the target region. Black dots indicate the positions of mutations (nucleotides 8 and 11).
FIG. 2.
Polioviruses resistant to siP. (A) Population sequencing of the viral siP region after one passage in siP-transfected cells. Arrows indicate newly arising mutations. (B) Sequences of individual viral genomes in the siP region appearing after 10 passages. Only nucleotides different from the wild-type sequence (shown on top) are spelled out. If a sequence was isolated more than once, then the number of isolations is given in the column second from the left. Six viral populations (numbered on the left as 1 to 6) were sequenced. Populations 1 to 3 are derived from passaging virus at an MOI of 10, and populations 4 to 6 are derived from passaging virus at an MOI of 0.1. Population pairs 1 and 4, 2 and 5, and 3 and 6 were related, as each pair was derived from one viral stock following two initial passages under siP. (C) Frequency of each mutation observed in siP-resistant viral population. Poliovirus was collected after 10 rounds of replication in siP-transfected HeLa cells. The target region was amplified by RT-PCR and cloned, and 70 clones were analyzed by sequencing. The mutation frequency was calculated as the number of times that a particular position was mutated divided by the total number of sequences analyzed.
FIG. 3.
Contribution of individual mutations to siP-resistance of poliovirus. (A) Sequences corresponding to the wild type (WT) and the siP escape mutants. (B) HeLa cells treated with either siP (white bars) or control siRNA (black bars) were infected at an MOI of 1. Virus was collected, and titers were determined after 10 h. siC infections were done in triplicate; error bars denote standard deviations.
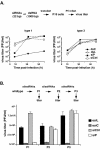
FIG. 4.
Targeting poliovirus with let-7 microRNA. (A) Schematic representation of recombinant poliovirus carrying let-7 sequences. Either the identical let-7 RNA sequence [Let-7 (+)] or the anti-sense let-7 RNA [Let-7 (−)] was inserted at position 703 of the poliovirus genome. (B) After transfection of viral RNA into HeLa cells, viral titers were determined by infectious focus assay. (C) Population sequencing of the region in which let-7 (−) or let-7 (+) sequences were inserted. The top sequence corresponds to the starting viral RNA before replicating in HeLa cells. Let-7 (−)/P1 (middle sequence) corresponds to a representative viral population after one passage in HeLa cells. Let-7 (+)/P1 (bottom) corresponds to Let-7 (+) virus after one passage in HeLa cells. Arrows indicate an A-to-G transition mutation arising in Let-7 (−) populations.
FIG. 5.
Relative escape strength of each mutation. (A) Schematic of the experiment. Two consecutive passages of a 1:1 mixture of the mutant viruses were performed with cells transfected with either siP or a control anti-rhinovirus siRNA. A change in the proportion of a given mutant after two passages was detected by RT-PCR and sequencing, as shown in Fig. 1D. Three siC mutants (B) and five siP mutants (C) were competed against each other. An increase in the proportion of a mutation in a given competition assay, defined as a winning trial, was scored as 1, a decrease was scored as 0, and no change in proportions was scored as 0.5. Each table (B and C) shows the scored mutants on the left, followed by their scores against each competing mutant (top). The fraction of winning trials (Fraction) was then calculated as the number of winning trials divided by the total number of trials. This allowed us to determine the relative escape efficiency of each mutation along the targeted region in the graphs shown on the right of panels B and C.
FIG. 6.
Strategy to prevent development of RNAi-resistant viruses. (A) Titers of type 1 and type 3 strains grown in P19 cells transfected with 1-kb-long dsRNA corresponding to the poliovirus type 1 genome (dsC), dsRNA to firefly luciferase (dsL), or siRNA siP or siCtrl. (B) Inhibition of poliovirus type 1 after consecutive passages in HeLa cells transfected with siRNAs siP and siCtrl or to esiRNAs prepared from dsC (esiC) and dsL (esiL). Viral titers are shown after each of three consecutive passages (P1, P2, and P3). For panels A and B, a scheme of the experiment is shown on top; see the text for details.
Similar articles
- Short interfering RNA confers intracellular antiviral immunity in human cells.
Gitlin L, Karelsky S, Andino R. Gitlin L, et al. Nature. 2002 Jul 25;418(6896):430-4. doi: 10.1038/nature00873. Epub 2002 Jun 26. Nature. 2002. PMID: 12087357 - Anti-coxsackieviral efficacy of RNA interference is highly dependent on genomic target selection and emergence of escape mutants.
Merl S, Wessely R. Merl S, et al. Oligonucleotides. 2007 Spring;17(1):44-53. doi: 10.1089/oli.2007.0057. Oligonucleotides. 2007. PMID: 17461762 - In Silico Design and Experimental Validation of siRNAs Targeting Conserved Regions of Multiple Hepatitis C Virus Genotypes.
ElHefnawi M, Kim T, Kamar MA, Min S, Hassan NM, El-Ahwany E, Kim H, Zada S, Amer M, Windisch MP. ElHefnawi M, et al. PLoS One. 2016 Jul 21;11(7):e0159211. doi: 10.1371/journal.pone.0159211. eCollection 2016. PLoS One. 2016. PMID: 27441640 Free PMC article. - Small interfering RNA (siRNA)-based therapeutic applications against viruses: principles, potential, and challenges.
Kang H, Ga YJ, Kim SH, Cho YH, Kim JW, Kim C, Yeh JY. Kang H, et al. J Biomed Sci. 2023 Oct 16;30(1):88. doi: 10.1186/s12929-023-00981-9. J Biomed Sci. 2023. PMID: 37845731 Free PMC article. Review. - RNAi therapy for HIV infection: principles and practicalities.
Bennasser Y, Yeung ML, Jeang KT. Bennasser Y, et al. BioDrugs. 2007;21(1):17-22. doi: 10.2165/00063030-200721010-00003. BioDrugs. 2007. PMID: 17263586 Free PMC article. Review.
Cited by
- The highly conserved 5' untranslated region as an effective target towards the inhibition of Enterovirus 71 replication by unmodified and appropriate 2'-modified siRNAs.
Deng JX, Nie XJ, Lei YF, Ma CF, Xu DL, Li B, Xu ZK, Zhang GC. Deng JX, et al. J Biomed Sci. 2012 Aug 13;19(1):73. doi: 10.1186/1423-0127-19-73. J Biomed Sci. 2012. PMID: 22889374 Free PMC article. - Application of RNA silencing to plant disease resistance.
Duan CG, Wang CH, Guo HS. Duan CG, et al. Silence. 2012 May 31;3(1):5. doi: 10.1186/1758-907X-3-5. Silence. 2012. PMID: 22650989 Free PMC article. - Inhibition of porcine circovirus type 2 replication in mice by RNA interference.
Liu J, Chen I, Chua H, Du Q, Kwang J. Liu J, et al. Virology. 2006 Apr 10;347(2):422-33. doi: 10.1016/j.virol.2005.12.006. Epub 2006 Jan 20. Virology. 2006. PMID: 16427679 Free PMC article. - RNA interference as a novel treatment strategy for chronic hepatitis B infection.
Hui RW, Mak LY, Seto WK, Yuen MF. Hui RW, et al. Clin Mol Hepatol. 2022 Jul;28(3):408-424. doi: 10.3350/cmh.2022.0012. Epub 2022 Feb 17. Clin Mol Hepatol. 2022. PMID: 35172540 Free PMC article. Review. - Coxsackievirus B3 and adenovirus infections of cardiac cells are efficiently inhibited by vector-mediated RNA interference targeting their common receptor.
Fechner H, Pinkert S, Wang X, Sipo I, Suckau L, Kurreck J, Dörner A, Sollerbrant K, Zeichhardt H, Grunert HP, Vetter R, Schultheiss HP, Poller W. Fechner H, et al. Gene Ther. 2007 Jun;14(12):960-71. doi: 10.1038/sj.gt.3302948. Epub 2007 Mar 22. Gene Ther. 2007. PMID: 17377597 Free PMC article.
References
- Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources