Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression - PubMed (original) (raw)
Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression
Marcela Covic et al. EMBO J. 2005.
Abstract
Nuclear factor kappaB (NF-kappaB) plays an important role in the transcriptional regulation of genes involved in inflammation and cell survival. Here, we show that coactivator-associated arginine methyltransferase CARM1/PRMT4 is a novel transcriptional coactivator of NF-kappaB and functions as a promoter-specific regulator of NF-kappaB recruitment to chromatin. Carm1 knockout cells showed impaired expression of a subset of NF-kappaB-dependent genes upon TNFalpha or LPS stimulation. CARM1 forms a complex with p300 and NF-kappaB in vivo and interacts directly with the NF-kappaB subunit p65 in vitro. CARM1 seems to act in a gene-specific manner mainly by enhancing NF-kappaB recruitment to cognate sites. Moreover, CARM1 synergistically coactivates NF-kappaB-mediated transactivation, in concert with the transcriptional coactivators p300/CREB-binding protein and the p160 family of steroid receptor coactivators. For at least a subset of CARM1-dependent NF-kappaB target genes, the enzymatic activities of both CARM1 and p300 are necessary for the observed synergy between CARM1 and p300. Our results suggest that the cooperative action between protein arginine methyltransferases and protein lysine acetyltransferases regulates NF-kappaB-dependent gene activation in vivo.
Figures
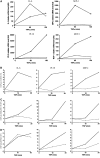
Figure 1
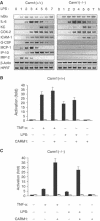
Impaired NF-κB-dependent gene expression in Carm1(−/−) cells in response to proinflammatory stimuli. (A) Impaired expression of MIP-2, MCP-1, G-CSF, ICAM1 and IP-10 in Carm1(−/−) MEF cells in response to LPS. Carm1(+/+) and Carm1(−/−) MEF cells were treated with LPS (10 μg/ml) and RNA isolated at the indicated time points, followed by RT–PCR determination of MIP-2, MCP-1, G-CSF, ICAM1, IP-10, IL-6, KC, COX-2, HPRT and β-actin mRNA. (B, C) Impaired NF-κB-dependent gene expression in Carm1(−/−) cells in response to TNFα and LPS. Carm1(+/+) MEF cells (B) and Carm1(−/−) MEF cells (C) were cotransfected with HIV-luc or HIVmut-κB-luc (2 μg) and pphRSVnt-β-gal (200 ng) together with CMV-CARM1 (500 ng) or CMV empty vector as indicated. Cells were subsequently stimulated with TNFα (10 ng/ml) or LPS (10 μg/ml) 24 h after transfection for 8 h. The indicated activation was determined by the ratio of the relative luciferase activity measured for the HIV-luc (black bars) or HIVmut-κB-luc (white bars) reporter gene after stimulation. The ratio obtained for untreated cells was arbitrarily set to 1. Error bars indicate s.e. of three independent experiments.
Figure 2
Expression levels of components of the NF-κB signaling pathway and nuclear–cytoplasmic shuttling of NF-κB is not affected in Carm1(−/−) MEFs. (A) Protein expression levels of p65, c-Rel, RelB, p300 and PARP-1 are not affected in Carm1(−/−) MEF cells. Carm1(+/+) and Carm1(−/−) MEF cells were treated with TNFα (10 ng/ml) for 20 min and cytoplasmic and nuclear extracts resolved by SDS–PAGE followed by subsequent immunoblot analysis for p65, c-Rel, RelB, p300 and PARP-1 (A, left and right panels). α-Tubulin was used as an internal standard. (B) mRNA expression levels of TLR4, TLR2 and IRAK4 are not impaired in Carm1(−/−) MEF cells. Carm1(+/+) and Carm1(−/−) MEF cells were treated with TNFα (10 ng/ml) (B, left and right panels) and RNA isolated at the indicated time points followed by RT–PCR analysis for TLR4, TLR2, IRAK4 and HPRT (B, left and right panels). (C) Degradation and re-synthesis of IκBα is not affected in Carm1(−/−) MEF cells. Carm1(+/+) and Carm1(−/−) MEF cells were treated with TNFα (10 ng/ml) (C, left and right panels) and whole-cell extracts isolated at the indicated time points and resolved by SDS–PAGE, followed by subsequent immunoblot analysis for IκBα. α-Tubulin was used as an internal standard. (D) Nuclear translocation of NF-κB is not delayed in Carm1(−/−) MEFs. Carm1(+/+) and Carm1(−/−) MEF cells were treated with TNFα (10 ng/ml) (D, upper and lower panels) and fixed with paraformaldehyde at the indicated time points followed by immunostaining for p65 and analysis by immunofluorescent microscopy.
Figure 3
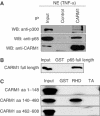
CARM1 forms a complex with NF-κB in vivo and directly binds to the NF-κB subunit p65 in vitro. (A) CARM1 and p65 form a complex in vivo. p65, CARM1 and p300 were coimmunoprecipitated (IP) in the presence of 120 mM NaCl from nuclear extract of TNFα-treated 293T cells (30 min, 10 ng/ml) using control IgGs and an anti-CARM1 antibody. Bound proteins were resolved by SDS–PAGE and subsequently detected by immunoblot (IB) analysis for p65, CARM1 and p300. Input lanes represent 10% of the input. (B) CARM1 directly binds to p65. Pull-down assays with purified GST and p65 full-length fused to GST (1 μg) and purified recombinant CARM1 full-length (0.1 μg) in the presence of 120 mM NaCl. Bound proteins were resolved by SDS–PAGE and detected by immunoblot analysis for CARM1. Input lanes represent 1% of the input. (C) The central domain of CARM1 binds to the RHD domain of p65. Pull-down assays with GST, the Rel-homology domain of p65 (RHD; aa 1–305) or the transactivation domain of p65 (TA; aa 441–551) fused to GST (1 μg) and different _in vitro_-translated CARM1 deletion mutants (N-terminal domain (aa 1–148), catalytic domain (aa 140–480) or transactivation domain (aa 462–608), respectively) in the presence of 120 mM NaCl. Bound proteins were resolved by SDS–PAGE visualized by autoradiography. Input lanes represent 1% of the input.
Figure 4
CARM1 synergistically coactivates the NF-κB-mediated transactivation, in concert with the transcriptional coactivator p300/CBP. (A, B) CARM1 synergistically activates together with p300 NF-κB-dependent gene expression in response to proinflammatory stimuli. Carm1(+/+) MEF cells (left panel) and Carm1(−/−) MEF cells (right panel) were cotransfected with HIV-luc or HIVmut-κB-luc (2 μg) and pphRSVnt-β-gal (200 ng), together with CMV-CARM1 (200 ng) and RSV-p300 (500 ng) or CMV and RSV empty vectors as indicated. Cells were subsequently stimulated with TNFα (10 ng/ml) (A) or LPS (10 μg/ml) (B) 24 h after transfection for 8 h. Cells were harvested 32 h after transfection and the indicated activation of NF-κB-dependent gene expression determined as described in Figure 1. (C) CARM1 synergistically activates together with p300 and p65-dependent gene expression. Carm1(+/+) MEF cells (left panel) and Carm1(−/−) MEF cells (right panel) were cotransfected with HIV-luc or HIVmut-κB-luc (2 μg) and pphRSVnt-β-gal (200 ng) together with CMV-p65 (80 ng) CMV-CARM1 (200 ng) and RSV-p300 (500 ng) or CMV and RSV empty vectors as indicated. The indicated activation of NF-κB-dependent gene expression was determined as described in Figure 1.
Figure 5
CARM1 synergistically coactivates NF-κB-dependent transcription in concert with the transcriptional coactivators p300/CBP and the p160 family of steroid receptor coactivators. (A, B) CARM1 synergistically activates together with p300 and SRC-2 p65-dependent gene expression. Carm1(+/+) MEF cells (A) and Carm1(−/−) MEF cells (B) were cotransfected with HIV-luc or HIVmut-κB-luc (2 μg) and pphRSVnt-β-gal (200 ng), together with CMV-p65 (80 ng), CMV-CARM1 (200 ng), CMV-SRC-2 (300 ng) and RSV-p300 (500 ng) or CMV and RSV empty vectors as indicated. Cells were harvested 32 h after transfection and the indicated activation of NF-κB-dependent gene expression determined as described in Figure 1. (C) CARM1 synergistically activates together with p300 and p65 NF-κB-dependent gene expression in COS-1 cells. COS-1 cells were cotransfected with HIV-luc or HIVmut-κB-luc (2 μg) and pphRSVnt-β-gal (200 ng) together with CMV-p65 (50 ng) CMV-CARM1 (200 ng), CMV-SRC-2 (300 ng) and RSV-p300 (500 ng) or CMV and RSV empty vectors as indicated. The indicated activation of NF-κB-dependent gene expression was determined as described in Figure 1.
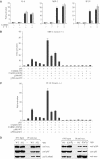
Figure 6
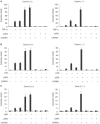
CARM1 functions in a gene-specific manner. (A) Quantification of TNFα-stimulated MIP-2, IP-10, MCP-1 and IL-6 transcription Carm1(+/+) MEF cells (filled circles) and Carm1(−/−) MEF cells (empty circles) by real-time PCR. (B) Quantitative ChIP assays with anti-CARM1 antibodies were performed for the indicated genes on TNFα-stimulated Carm1(+/+) (filled circles) and Carm1(−/−) MEF cells (empty circles). (C) Anti-methyl-H3(R17) ChIP assay for the indicated genes on TNFα-stimulated Carm1(+/+) (filled circles) and Carm1(−/−) MEF cells (empty circles). (D) ChIP assays with anti-p65 antibodies were performed for the indicated genes on TNFα-stimulated Carm1(+/+) (filled circles) and Carm1(−/−) MEF cells (empty circles).
Figure 7
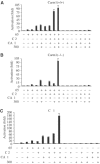
Enzymatic activities of CARM1 and p300/CBP are required for full cooperativity between CARM1 and p300. (A) Carm1(+/+) MEF cells (black bars) and Carm1(−/−) MEF cells (dashed bars) were cotransfected with pGL3-MIP-2(−770), pGL3-IP-10(−533) or pOLUC-IL-6(−303/+23) (5 μg) and pphRSVnt-β-gal (200 ng) alone or together with CMV-CARM1 (200 ng) or CMV empty vectors as indicated. Cells were subsequently stimulated with TNFα (10 ng/ml) for 6 h. Cells were harvested 32 h after transfection and the indicated activation of NF-κB-dependent gene expression determined as described in Figure 1. (B, C) Enzymatic activities of CARM1 and p300/CBP are required for full cooperativity between CARM1 and p300. Carm1(−/−) MEF cells were cotransfected with pGL3-MIP-2(−770) (B) or pGL3-IP-10(−533) (C) (5 μg) and pphRSVnt-β-gal (200 ng), together with CMV-p65 (80 ng) CMV-CARM1-WT or CARM1-E267Q (200 ng), CMV-p300-WT or p300mutAT2 (750 ng) and CMV empty vectors as indicated. Cells were harvested 32 h after transfection and the indicated activation of NF-κB-dependent gene expression determined as described in Figure 1. (D) Both wild type and mutant forms of CARM1 and p300 interact to the same extent with p65 in vivo. 293T cells were transfected with CMV expression vectors, either for wild type or enzymatic mutant forms of myc tagged p300 (left panel) or CARM1 (right panel) as well as pphCMV-kozak-myc empty vector. p65, CARM1 and p300 were coimmunoprecipitated (IP) from nuclear extract of TNFα-treated 293T cells (30 min, 10 ng/ml) using an anti-myc antibody. Bound proteins were resolved by SDS–PAGE and subsequently detected by immunoblot (IB) analysis for p65, CARM1 and p300. Input lanes represent 10% of the input.
Similar articles
- Protein arginine methyltransferase 1 coactivates NF-kappaB-dependent gene expression synergistically with CARM1 and PARP1.
Hassa PO, Covic M, Bedford MT, Hottiger MO. Hassa PO, et al. J Mol Biol. 2008 Mar 28;377(3):668-78. doi: 10.1016/j.jmb.2008.01.044. Epub 2008 Jan 26. J Mol Biol. 2008. PMID: 18280497 - Coactivator-associated arginine methyltransferase-1 enhances nuclear factor-kappaB-mediated gene transcription through methylation of histone H3 at arginine 17.
Miao F, Li S, Chavez V, Lanting L, Natarajan R. Miao F, et al. Mol Endocrinol. 2006 Jul;20(7):1562-73. doi: 10.1210/me.2005-0365. Epub 2006 Feb 23. Mol Endocrinol. 2006. PMID: 16497732 - The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators.
Stallcup MR, Kim JH, Teyssier C, Lee YH, Ma H, Chen D. Stallcup MR, et al. J Steroid Biochem Mol Biol. 2003 Jun;85(2-5):139-45. doi: 10.1016/s0960-0760(03)00222-x. J Steroid Biochem Mol Biol. 2003. PMID: 12943698 Review. - Bile salt excretory pump: biology and pathobiology.
Suchy FJ, Ananthanarayanan M. Suchy FJ, et al. J Pediatr Gastroenterol Nutr. 2006 Jul;43 Suppl 1:S10-6. doi: 10.1097/01.mpg.0000226385.71859.5f. J Pediatr Gastroenterol Nutr. 2006. PMID: 16819395 Review.
Cited by
- CNNArginineMe: A CNN structure for training models for predicting arginine methylation sites based on the One-Hot encoding of peptide sequence.
Zhao J, Jiang H, Zou G, Lin Q, Wang Q, Liu J, Ma L. Zhao J, et al. Front Genet. 2022 Oct 17;13:1036862. doi: 10.3389/fgene.2022.1036862. eCollection 2022. Front Genet. 2022. PMID: 36324513 Free PMC article. - The Role of Protein Methyltransferases in Immunity.
Song C, Kim MY, Cho JY. Song C, et al. Molecules. 2024 Jan 11;29(2):360. doi: 10.3390/molecules29020360. Molecules. 2024. PMID: 38257273 Free PMC article. Review. - CCL20 is elevated during obesity and differentially regulated by NF-κB subunits in pancreatic β-cells.
Burke SJ, Karlstad MD, Regal KM, Sparer TE, Lu D, Elks CM, Grant RW, Stephens JM, Burk DH, Collier JJ. Burke SJ, et al. Biochim Biophys Acta. 2015 Jun;1849(6):637-52. doi: 10.1016/j.bbagrm.2015.03.007. Epub 2015 Apr 13. Biochim Biophys Acta. 2015. PMID: 25882704 Free PMC article. - Mediator complex interaction partners organize the transcriptional network that defines neural stem cells.
Quevedo M, Meert L, Dekker MR, Dekkers DHW, Brandsma JH, van den Berg DLC, Ozgür Z, van IJcken WFJ, Demmers J, Fornerod M, Poot RA. Quevedo M, et al. Nat Commun. 2019 Jun 17;10(1):2669. doi: 10.1038/s41467-019-10502-8. Nat Commun. 2019. PMID: 31209209 Free PMC article. - The CARM1-p300-c-Myc-Max (CPCM) transcriptional complex regulates the expression of CUL4A/4B and affects the stability of CRL4 E3 ligases in colorectal cancer.
Lu W, Yang C, He H, Liu H. Lu W, et al. Int J Biol Sci. 2020 Feb 4;16(6):1071-1085. doi: 10.7150/ijbs.41230. eCollection 2020. Int J Biol Sci. 2020. PMID: 32140074 Free PMC article.
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D (2000) Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103: 667–678 - PubMed
- An W, Kim J, Roeder RG (2004) Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117: 735–748 - PubMed
- Baeuerle PA, Baltimore D (1988) I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science 242: 540–546 - PubMed
- Baldwin AS Jr (1996) The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol 14: 649–683 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK062248/DK/NIDDK NIH HHS/United States
- R56 DK062248/DK/NIDDK NIH HHS/United States
- DK62248/DK/NIDDK NIH HHS/United States
- P30 ES007784/ES/NIEHS NIH HHS/United States
- ES07784/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous