Modulating HIV-1 replication by RNA interference directed against human transcription elongation factor SPT5 - PubMed (original) (raw)
Modulating HIV-1 replication by RNA interference directed against human transcription elongation factor SPT5
Yueh-Hsin Ping et al. Retrovirology. 2004.
Abstract
Background: Several cellular positive and negative elongation factors are involved in regulating RNA polymerase II processivity during transcription elongation in human cells. In recruiting several of these regulatory factors to the 5' long terminal repeat (LTR) promoter during transcription elongation, HIV-1 modulates replication of its genome in a process mediated by the virus-encoded transactivator Tat. One particular cellular regulatory factor, DSIF subunit human SPT5 (hSpt5), has been implicated in both positively and negatively regulating transcriptional elongation but its role in Tat transactivation in vivo and in HIV-1 replication has not been completely elucidated.
Results: To understand the in vivo function of hSpt5 and define its role in Tat transactivation and HIV-1 replication, we used RNA interference (RNAi) to specifically knockdown hSpt5 expression by degrading hSpt5 mRNA. Short-interfering RNA (siRNA) designed to target hSpt5 for RNAi successfully resulted in knockdown of both hSpt5 mRNA and protein levels, and did not significantly affect cell viability. In contrast to hSpt5 knockdown, siRNA-mediated silencing of human mRNA capping enzyme, a functionally important hSpt5-interacting cellular protein, was lethal and showed a significant increase in cell death over the course of the knockdown experiment. In addition, hSpt5 knockdown led to significant decreases in Tat transactivation and inhibited HIV-1 replication, indicating that hSpt5 was required for mediating Tat transactivation and HIV-1 replication.
Conclusions: The findings presented here showed that hSpt5 is a bona fide positive regulator of Tat transactivation and HIV-1 replication in vivo. These results also suggest that hSpt5 function in transcription regulation and mRNA capping is essential for a subset of cellular and viral genes and may not be required for global gene expression.
Figures
Figure 1
Specific silencing of hSpt5 expression by RNAi. (A) hSpt5 mRNA is 3261 nucleotides in length. siRNA targeting sequence for hSpt5 was selected from position 407 to 427 relative to the start codon. As a specific control, mutant siRNA containing 2 nucleotide mismatches (underline) between the target mRNA and the antisense of siRNA at the hypothetical cleavage sites of the mRNA was generated. (B) Evaluation of specific hSpt5 siRNA activity by RT-PCR. Total cellular mRNA was prepared from HeLa cells transfected without siRNA or with hSpt5 duplex or control siRNAs and was followed by RT-PCR, as described in Material and Methods. Each RT-PCR reaction included 100 ng total cellular mRNA, gene-specific primer sets for hSpt5 and hCycT1 amplification (0.5 μM for each primer), 200 μM dNTP, 1.2 mM MgSO4 and 1U of RT/platinum Taq mix. Primer sets for hSpt5 produced 2.6 kb products while hCycT1 produced 1.8 kb products. RT-PCR products were resolved on a 1% agarose gel and viewed by ethidium bromide staining. RT-PCR products are shown from cells that were not transfected with siRNA (lane 1), or cells transfected with single-stranded antisense hSpt5 siRNA (hSpt5 (AS), lane 2), hSpt5 duplex siRNA (hSpt5 (DS), lane 3), or mismatch hSpt5 duplex siRNA (hSpt5-mm (DS), lane 4). Lane M is a marker lane. (C) Analysis of specific hSpt5 siRNA activity by western blotting. Cell lysates were prepared from HeLa cells mock-transfected without siRNA (lane 1), or transfected with single-stranded antisense hSpt5 siRNA (hSpt5 (AS), lane 2), hSpt5 duplex siRNA (hSpt5 (DS), lane 3), or mismatch hSpt5 duplex siRNA (hSpt5-mm (DS), lane 4). Cell lysates were analyzed by 10% SDS-PAGE. Protein contents were detected by immunoblotting assay with antibodies against hSpt5 (top panel) and hCycT1 (lower panel).
Figure 2
Kinetics of specific hSpt5 siRNA activity by Western blotting. HeLa cells were transfected with single-stranded antisense hSpt5 siRNA (hSpt5 (AS), lanes 1–7), hSpt5 duplex siRNA (hSpt5 (DS), lanes 8–14), or mismatch hSpt5 duplex siRNA (hSpt5-mm (DS), lanes 15–21) having 2 nucleotide mismatches between the target mRNA and the antisense strand of siRNA at the hypothetical cleavage site of the mRNA. Cells were harvested at various times post transfection. Protein content was resolved on 10% SDS-PAGE, transferred onto PVDF membranes, and immunoblotted with antibodies against hSpt5 (top bands) and hCycT1 as an internal control (lower bands).
Figure 3
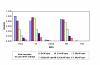
Analysis of cell viability by counting trypan blue-stained cells. HeLa cells were transfected with Lipofectamine with various siRNAs or no siRNA. Three siRNA duplexes, including hSpt5 siRNA (yellow), mismatch hSpt5 siRNA (light blue) and siRNA targeting human capping enzyme (HCE, red), were used in these experiments. Controls for viability included cells mock-transfected with no siRNA (dark blue) or cells transfected with single-stranded antisense hSpt5 siRNA (purple). At various times after transfection, cells floating in the medium were collected and counted in the presence of 0.2% trypan blue. Cells that took up dye (stained blue) were counted as not viable.
Figure 4
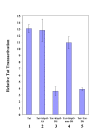
Effect of hSpt5 siRNA on HIV-1 Tat transactivation in Magi cells. Quantified effect of siRNA on HIV-1 Tat transactivation was determined by measuring β-galactosidase activity. Magi cells were co-transfected with pTat-RFP plasmid and various siRNAs targeting hSpt5 or Tat and harvested at 48 h post-transfection. Activity of β-galactosidase was measured using the β-Galactosidase Enzyme Assay System (Promega). Tat transactivation was determined by the ratio of β-galactosidase activity in pTat-RFP transfected cells to activity measured in cells without pTat-RFP. The inhibitory effect of siRNA was determined by normalizing Tat transactivation activity to the amount of Tat-RFP protein. Tat transactivation was measured for Magi cells transfected with pTat-RFP only (lane 1), or Tat-RFP transfected with single-stranded antisense hSpt5 siRNA (hSpt5-AS, lane 2), hSpt5 duplex siRNA (hSpt5-DS, lane 3), mismatch hSpt5 duplex siRNA (hSpt5-mm-DS, lane 4), or Tat siRNA duplex (Tat-DS, lane 5). Results are representative of three independent experiments.
Figure 5
siRNA targeting hSpt5 modulate HIV-1 replication. HeLa-CD4-LTR/β-galactosidase (Magi) cells were mock-transfected (mock), or transfected with single-stranded antisense hSpt5 siRNA (AS), hSpt5 duplex siRNA (siRNA), mismatched hSpt5 duplex siRNA (MM) or Vif duplex siRNA (T98). 24 h after the first transfection, a second siRNA transfection was performed. 24 h later, cells were infected with HIVNL-GFP, an infectious molecular clone of HIV-1. Cells infected with virus and not treated with oligofectamine are shown (mock). HIV-1 Tat-mediated transactivation of the 5' LTR led to β-galactosidase production, which was quantified 48 h post-infection. Cells treated with duplex siRNA targeting Vif (lanes marked T98 [47]) served as a positive control. Serial double dilutions of the viral inoculum (in cpm of RT activity) are consistent with 32-fold decreases in viral replication.
Figure 6
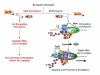
Model for Tat transactivation in absence or presence of SPT5. See text for details.
Similar articles
- Inhibition of human immunodeficiency virus type 1 replication by RNA interference directed against human transcription elongation factor P-TEFb (CDK9/CyclinT1).
Chiu YL, Cao H, Jacque JM, Stevenson M, Rana TM. Chiu YL, et al. J Virol. 2004 Mar;78(5):2517-29. doi: 10.1128/jvi.78.5.2517-2529.2004. J Virol. 2004. PMID: 14963154 Free PMC article. - Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences.
Bourgeois CF, Kim YK, Churcher MJ, West MJ, Karn J. Bourgeois CF, et al. Mol Cell Biol. 2002 Feb;22(4):1079-93. doi: 10.1128/MCB.22.4.1079-1093.2002. Mol Cell Biol. 2002. PMID: 11809800 Free PMC article. - CRISPRi-mediated depletion of Spt4 and Spt5 reveals a role for DSIF in the control of HIV latency.
Krasnopolsky S, Novikov A, Kuzmina A, Taube R. Krasnopolsky S, et al. Biochim Biophys Acta Gene Regul Mech. 2021 Jan;1864(1):194656. doi: 10.1016/j.bbagrm.2020.194656. Epub 2020 Dec 15. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 33333262 - Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator.
Marcello A, Zoppé M, Giacca M. Marcello A, et al. IUBMB Life. 2001 Mar;51(3):175-81. doi: 10.1080/152165401753544241. IUBMB Life. 2001. PMID: 11547919 Review. - Regulatory functions of Cdk9 and of cyclin T1 in HIV tat transactivation pathway gene expression.
Romano G, Kasten M, De Falco G, Micheli P, Khalili K, Giordano A. Romano G, et al. J Cell Biochem. 1999 Dec 1;75(3):357-68. J Cell Biochem. 1999. PMID: 10536359 Review.
Cited by
- DSIF contributes to transcriptional activation by DNA-binding activators by preventing pausing during transcription elongation.
Zhu W, Wada T, Okabe S, Taneda T, Yamaguchi Y, Handa H. Zhu W, et al. Nucleic Acids Res. 2007;35(12):4064-75. doi: 10.1093/nar/gkm430. Epub 2007 Jun 12. Nucleic Acids Res. 2007. PMID: 17567605 Free PMC article. - RNA interference for antiviral therapy.
Ketzinel-Gilad M, Shaul Y, Galun E. Ketzinel-Gilad M, et al. J Gene Med. 2006 Aug;8(8):933-50. doi: 10.1002/jgm.929. J Gene Med. 2006. PMID: 16779870 Free PMC article. Review. - Mechanisms of HIV Transcriptional Regulation and Their Contribution to Latency.
Schiralli Lester GM, Henderson AJ. Schiralli Lester GM, et al. Mol Biol Int. 2012;2012:614120. doi: 10.1155/2012/614120. Epub 2012 Jun 3. Mol Biol Int. 2012. PMID: 22701796 Free PMC article. - Biochemical and genetic analysis of RNA cap guanine-N2 methyltransferases from Giardia lamblia and Schizosaccharomyces pombe.
Hausmann S, Ramirez A, Schneider S, Schwer B, Shuman S. Hausmann S, et al. Nucleic Acids Res. 2007;35(5):1411-20. doi: 10.1093/nar/gkl1150. Epub 2007 Feb 6. Nucleic Acids Res. 2007. PMID: 17284461 Free PMC article. - Coxsackievirus B3 and adenovirus infections of cardiac cells are efficiently inhibited by vector-mediated RNA interference targeting their common receptor.
Fechner H, Pinkert S, Wang X, Sipo I, Suckau L, Kurreck J, Dörner A, Sollerbrant K, Zeichhardt H, Grunert HP, Vetter R, Schultheiss HP, Poller W. Fechner H, et al. Gene Ther. 2007 Jun;14(12):960-71. doi: 10.1038/sj.gt.3302948. Epub 2007 Mar 22. Gene Ther. 2007. PMID: 17377597 Free PMC article.
References
- Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6:2201–2213. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI043198/AI/NIAID NIH HHS/United States
- R33 AI088595/AI/NIAID NIH HHS/United States
- AI43198/AI/NIAID NIH HHS/United States
- R01 AI032890/AI/NIAID NIH HHS/United States
- RR11489/RR/NCRR NIH HHS/United States
- AI32890/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources