Contrasting effects of natural selection on human and chimpanzee CC chemokine receptor 5 - PubMed (original) (raw)
Comparative Study
. 2005 Feb;76(2):291-301.
doi: 10.1086/427927. Epub 2004 Dec 29.
Affiliations
- PMID: 15625621
- PMCID: PMC1196374
- DOI: 10.1086/427927
Comparative Study
Contrasting effects of natural selection on human and chimpanzee CC chemokine receptor 5
Stephen Wooding et al. Am J Hum Genet. 2005 Feb.
Erratum in
- Am J Hum Genet. 2005 Apr;76(4):715
Abstract
Human immunodeficiency virus type 1 (HIV-1) evolved via cross-species transmission of simian immunodeficiency virus (SIVcpz) from chimpanzees (Pan troglodytes). Chimpanzees, like humans, are susceptible to infection by HIV-1. However, unlike humans, infected chimpanzees seldom develop immunodeficiency when infected with SIVcpz or HIV-1. SIVcpz and most strains of HIV-1 require the cell-surface receptor CC chemokine receptor 5 (CCR5) to infect specific leukocyte subsets, and, subsequent to infection, the level of CCR5 expression influences the amount of HIV-1 entry and the rate of HIV-1 replication. Evidence that variants in the 5' cis-regulatory region of CCR5 (5'CCR5) affect disease progression in humans suggests that variation in CCR5 might also influence the response of chimpanzees to HIV-1/SIVcpz. To determine whether patterns of genetic variation at 5'CCR5 in chimpanzees are similar to those in humans, we analyzed patterns of DNA sequence variation in 37 wild-born chimpanzees (26 P. t. verus, 9 P. t. troglodytes, and 2 P. t. schweinfurthii), along with previously published 5'CCR5 data from 112 humans and 50 noncoding regions in the human and chimpanzee genomes. These analyses revealed that patterns of variation in 5'CCR5 differ dramatically between chimpanzees and humans. In chimpanzees, 5'CCR5 was less diverse than 80% of noncoding regions and was characterized by an excess of rare variants. In humans, 5'CCR5 was more diverse than 90% of noncoding regions and had an excess of common variants. Under a wide range of demographic histories, these patterns suggest that, whereas human 5'CCR5 has been subject to balancing selection, chimpanzee 5'CCR5 has been influenced by a selective sweep. This result suggests that chimpanzee 5'CCR5 might harbor or be linked to functional variants that influence chimpanzee resistance to disease caused by SIVcpz/HIV-1.
Figures
Figure 1
Geographical distributions of chimpanzee subspecies in Africa (redrawn from the article by Gagneux et al. [2001])
Figure 2
CCR5 variation in chimpanzees and humans. A, Schematic drawing (not to scale) of CCR5, including exons (boxes) and introns. Polymorphisms found in the 5′ _cis_-regulatory region of CCR5, which spans from −2867 to −1745, are indicated and numbered. Genomic sequencing revealed 15 and 10 polymorphisms in humans (bottom) and chimpanzees (top), respectively. An excessive number of singletons is found in chimpanzees (asterisks [*]), whereas humans have more intermediate-frequency variants than expected under neutrality. B, 5′CCR5 haplotypes observed in chimpanzees. Positions are shown relative to the CCR5 translational start site (+1). Only positions that are variable in chimpanzees or that differ between the chimpanzee sequence and the human consensus sequence are shown. Columns at right indicate the number of times each haplotype was observed in P. t. verus (Ptv), P. t. troglodytes (Ptt), P. t. schweinfurthii (Pts), and the total sample. Hs = the human consensus sequence. C, Two of the CCR5 mRNA transcripts found in humans. An A→G mutation at −2205 in the splice-acceptor site of exon 2A eliminates the production of CCR5A in most chimpanzees. This site has reverted to A in some P. t. troglodytes, suggesting that they have regained the capability to make CCR5A.
Figure 3
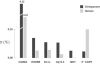
Nucleotide diversity in chimpanzees and humans. Each pair of bars represents a different locus/region. Data are from Stone et al. (2002) (for mtDNA), Deinard and Kidd (1999) (for HOXB6), Yu et al. (2002, 2003) (for 50 nc), Kaessmann et al. (1999, 2001) (for Xq13.3), Stone et al. (2002) (for NRY [noncoding region of the Y chromosome]), and from the present study (5′CCR5). The 5′CCR5 was the only region for which humans are more diverse than chimpanzees. In addition, whereas diversity at 5′CCR5 in chimpanzees was lower than that in the 50 noncoding (nc) regions (Yu et al. 2002, 2003), diversity at 5′CCR5 in humans was higher than that in the 50 noncoding regions.
Figure 4
MS networks of haplotypes of 5′CCR5 in chimpanzees (A) and humans (B). Each circle represents a different haplotype, and the size of each circle is proportional to the relative frequency of the haplotype. For each haplotype, the extent of shading indicates the fraction of observations in Africans, Asians, Europeans, or different subspecies of chimpanzee. The lines between haplotypes correspond to one nucleotide substitution, except where the number of nucleotides is indicated in parentheses. Reticulations indicate ambiguous relationships.
Figure 5
CIs for tests of Tajima’s D, estimated for 5′CCR5 in chimpanzees (A) and humans (B). Each CI shows the demographic parameters for which neutrality could be rejected (red) or not rejected (blue) on the basis of Tajima’s D test. On the axes, N 1 is the effective population size of the ancestral population, τ is the time of expansion (in generations), and N 0 is the effective population size of the population after expansion. For example, if it is assumed that the chimpanzee population has a generation time of 20 years, that the ancient population size was 25,000, and that the population grew 250-fold 100,000 years ago, then
τ=2(100,000/20)/25,000=0.4
, and the hypothesis of neutrality cannot be rejected. For the negative value of Tajima’s D in chimpanzee 5′CCR5, the hypothesis of neutrality is rejected under constant population size or recent growth but not under ancient growth. In humans, the value of Tajima’s D for 5′CCR5 is positive, and neutrality cannot be rejected under either constant population size or recent population growth. Neutrality is rejected under older population growth.
Similar articles
- The epidemiology of simian immunodeficiency virus infection in a large number of wild- and captive-born chimpanzees: evidence for a recent introduction following chimpanzee divergence.
Switzer WM, Parekh B, Shanmugam V, Bhullar V, Phillips S, Ely JJ, Heneine W. Switzer WM, et al. AIDS Res Hum Retroviruses. 2005 May;21(5):335-42. doi: 10.1089/aid.2005.21.335. AIDS Res Hum Retroviruses. 2005. PMID: 15929695 - Neutralization properties of simian immunodeficiency viruses infecting chimpanzees and gorillas.
Barbian HJ, Decker JM, Bibollet-Ruche F, Galimidi RP, West AP Jr, Learn GH, Parrish NF, Iyer SS, Li Y, Pace CS, Song R, Huang Y, Denny TN, Mouquet H, Martin L, Acharya P, Zhang B, Kwong PD, Mascola JR, Verrips CT, Strokappe NM, Rutten L, McCoy LE, Weiss RA, Brown CS, Jackson R, Silvestri G, Connors M, Burton DR, Shaw GM, Nussenzweig MC, Bjorkman PJ, Ho DD, Farzan M, Hahn BH. Barbian HJ, et al. mBio. 2015 Apr 21;6(2):e00296-15. doi: 10.1128/mBio.00296-15. mBio. 2015. PMID: 25900654 Free PMC article. - Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes.
Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. Gao F, et al. Nature. 1999 Feb 4;397(6718):436-41. doi: 10.1038/17130. Nature. 1999. PMID: 9989410 - [SIV as a source of HIV. On the origin of human immunodeficiency viruses from non-human primates].
Gürtler L. Gürtler L. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2004 Jul;47(7):680-4. doi: 10.1007/s00103-004-0862-z. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2004. PMID: 15254823 Review. German. - The HIV-1 pandemic: does the selective sweep in chimpanzees mirror humankind's future?
de Groot NG, Bontrop RE. de Groot NG, et al. Retrovirology. 2013 May 24;10:53. doi: 10.1186/1742-4690-10-53. Retrovirology. 2013. PMID: 23705941 Free PMC article. Review.
Cited by
- Genetic adaptations to SIV across chimpanzee populations.
Pawar H, Ostridge HJ, Schmidt JM, Andrés AM. Pawar H, et al. PLoS Genet. 2022 Aug 25;18(8):e1010337. doi: 10.1371/journal.pgen.1010337. eCollection 2022 Aug. PLoS Genet. 2022. PMID: 36007015 Free PMC article. - Role of CCL3L1-CCR5 genotypes in the epidemic spread of HIV-1 and evaluation of vaccine efficacy.
Kulkarni H, Marconi VC, Agan BK, McArthur C, Crawford G, Clark RA, Dolan MJ, Ahuja SK. Kulkarni H, et al. PLoS One. 2008;3(11):e3671. doi: 10.1371/journal.pone.0003671. Epub 2008 Nov 7. PLoS One. 2008. PMID: 18989363 Free PMC article. - Intronic deletions that disrupt mRNA splicing of the tva receptor gene result in decreased susceptibility to infection by avian sarcoma and leukosis virus subgroup A.
Reinišová M, Plachý J, Trejbalová K, Šenigl F, Kučerová D, Geryk J, Svoboda J, Hejnar J. Reinišová M, et al. J Virol. 2012 Feb;86(4):2021-30. doi: 10.1128/JVI.05771-11. Epub 2011 Dec 14. J Virol. 2012. PMID: 22171251 Free PMC article. - Different selective pressures shape the molecular evolution of color vision in chimpanzee and human populations.
Verrelli BC, Lewis CM Jr, Stone AC, Perry GH. Verrelli BC, et al. Mol Biol Evol. 2008 Dec;25(12):2735-43. doi: 10.1093/molbev/msn220. Epub 2008 Oct 1. Mol Biol Evol. 2008. PMID: 18832077 Free PMC article. - Effective activation alleviates the replication block of CCR5-tropic HIV-1 in chimpanzee CD4+ lymphocytes.
Decker JM, Zammit KP, Easlick JL, Santiago ML, Bonenberger D, Hahn BH, Kutsch O, Bibollet-Ruche F. Decker JM, et al. Virology. 2009 Nov 10;394(1):109-18. doi: 10.1016/j.virol.2009.08.027. Epub 2009 Sep 12. Virology. 2009. PMID: 19748647 Free PMC article.
References
Electronic-Database Information
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human 5′CCR5 [accession numbers AF031236 and AF031237])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CCR5) - PubMed
References
- Alkhatib G, Combadiere C, Broder CC, Fent Y, Kennedy PE, Murphy PM, Berger EA (1996) CC CKR5: a RANTES, MIP-1π, MIP-1α receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955–1958 - PubMed
- Bamshad M, Wooding S (2003) Signatures of natural selection in the human genome. Nat Rev Genet 4:99–111 - PubMed
- Corbet S, Muller-Trutwin MC, Versmisse P, Delarue S, Ayouba A, Lewis J, Brunak S, Martin P, Brun-Vezinet F, Simon F, Barre-Sinoussi F, Mauclere P (2000) env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group n from the same geographic area. J Virol 74:529–534 - PMC - PubMed
- Cunningham AL, Li S, Juarez J, Lynch G, Alali M, Naif H (2000) The level of HIV-1 infection of macrophages is determined by interaction of viral and host cell genotypes. J Leukoc Biol 68:311–317 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI43279/AI/NIAID NIH HHS/United States
- R37 AI046326/AI/NIAID NIH HHS/United States
- ES-10058/ES/NIEHS NIH HHS/United States
- R21 AI046326/AI/NIAID NIH HHS/United States
- R01 AI043279/AI/NIAID NIH HHS/United States
- F32 ES012125/ES/NIEHS NIH HHS/United States
- R01 AI046326/AI/NIAID NIH HHS/United States
- RR-00064/RR/NCRR NIH HHS/United States
- R01 GM059290/GM/NIGMS NIH HHS/United States
- M01 RR000064/RR/NCRR NIH HHS/United States
- GM-59290/GM/NIGMS NIH HHS/United States
- ES-12125/ES/NIEHS NIH HHS/United States
- AI46326/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources