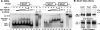The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members - PubMed (original) (raw)
The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members
William M Mahoney Jr et al. Biochem J. 2005.
Abstract
Members of the highly related TEF-1 (transcriptional enhancer factor-1) family (also known as TEAD, for TEF-1, TEC1, ABAA domain) bind to MCAT (muscle C, A and T sites) and A/T-rich sites in promoters active in cardiac, skeletal and smooth muscle, placenta, and neural crest. TEF-1 activity is regulated by interactions with transcriptional co-factors [p160, TONDU (Vgl-1, Vestigial-like protein-1), Vgl-2 and YAP65 (Yes-associated protein 65 kDa)]. The strong transcriptional co-activator YAP65 interacts with all TEF-1 family members, and, since YAP65 is related to TAZ (transcriptional co-activator with PDZ-binding motif), we wanted to determine if TAZ also interacts with members of the TEF-1 family. In the present study, we show by GST (glutathione S-transferase) pull-down assays, by co-immunoprecipitation and by modified mammalian two-hybrid assays that TEF-1 interacts with TAZ in vitro and in vivo. Electrophoretic mobility-shift assays with purified TEF-1 and GST-TAZ fusion protein showed that TAZ interacts with TEF-1 bound to MCAT DNA. TAZ can interact with endogenous TEF-1 proteins, since exogenous TAZ activated MCAT-dependent reporter promoters. Like YAP65, TAZ interacted with all four TEF-1 family members. GST pull-down assays with increasing amounts of [35S]TEF-1 and [35S]RTEF-1 (related TEF-1) showed that TAZ interacts more efficiently with TEF-1 than with RTEF-1. This differential interaction also extended to the interaction of TEF-1 and RTEF-1 with TAZ in vivo, as assayed by a modified mammalian two-hybrid experiment. These data show that differential association of TEF-1 proteins with transcriptional co-activators may regulate the activity of TEF-1 family members.
Figures
Figure 1. TEF-1, YAP65 and TAZ proteins
(A) The TEF-1 family of proteins has a number of conserved domains: the DNA-binding TEA domain (DBD), the proline-rich domain (PRO) and the C-terminal half of TEF-1. The region of TEF-1 that interacts with YAP65 is shown. Additionally, the amino acid identity between rTEF-1 and rRTEF-1 is shown for each domain. (B) TAZ is homologous with YAP65. The diagram shows the structure of YAP65 and TAZ. The following domains are highlighted: the WW (two conserved tryptophan residues spaced 20–22 amino acids apart) domains, the CC (coiled-coil) domains, the PDZ-interaction domain, the TEF-1-interaction domain, and the specific amino acid which regulates interaction with 14-3-3 (Ser89 in murine TAZ and Ser112 in mouse YAP65). (C) Homology of YAP65 and TAZ. Sequence alignment of the mouse TAZ and mouse YAP65 proteins within the region of YAP65 that interacts with the TEF-1 family of proteins [24]. Identical amino acids are boxed and shaded, and similar amino acids are boxed. *, 14-3-3-interaction motif: R-S-X-pS-X-P. (D) TAZ and YAP65 are expressed in muscle tissues. Left: Western blots on nuclear extracts (20 μg) from C2C12 myoblasts and myotubes were performed with the anti-YAP65/TAZ antibody (αYAP/TAZ). The immunoreactive bands are specific, as they co-migrate with overexpressed TAZ-N–FLAG and HA–YAP65 and pre-incubation of the antibody (anti-FLAG, α-FLAG, or anti-HA, α-HA) with immunogenic peptide resulted in no immunoreactivity (results not shown). Right: RT-PCR analyses on RNA from adult mouse heart and skeletal muscle (leg) were carried out with primers for YAP65 and TAZ. Both YAP65 and TAZ are expressed. Controls for these reactions, no RNA and no RT (−RNA −RT), resulted in no DNA products. Sizes in kDa (left) or kb (right) are indicated.
Figure 2. TEF-1 specifically interacts with the N-terminus of TAZ in vitro
(A) Interaction between TEF-1 and TAZ was assayed by GST pull-down assays. GST-fusion proteins of the N-terminus (N; amino acids 1–239) and the C-terminus (C; amino acids 239–395) of TAZ were produced and purified. Both empty GST protein (G) and GST–agarose beads alone (−) were used as controls. These proteins (5 μg) were mixed with the following in vitro labelled [35S]methionine TEF-1 isoforms: TEF-1ε, TEF-1η, TEF-1εΔA, TEF-1ζ, TEF-1-D and TEF-1εΔD [see (B) for diagram]. The TEF-1 and TAZ proteins were mixed with interaction buffer and GST–agarose beads overnight at 4 °C. Protein complexes were washed extensively and separated by SDS/PAGE (12.5% gel). Lanes 1–6 represent 7% of the total volume of lysate probed in each reaction. TEF-1εΔD was produced at a lower level, so this portion of the gel is a longer exposure. (B) Schematic representation of TEF-1 isoforms utilized in the GST pull-down assay (left) and summary of the TAZ–TEF-1 interaction assay (right).
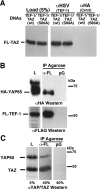
Figure 3. TEF-1 interacts with TAZ in vivo
(A) The interaction between TEF-1 and TAZ was confirmed by an in vivo Co-IP assay. H1299 cells were co-transfected with expression vectors for both TEF-1ε–HSV (XJ40-TEF-1ε-HSV) and FLAG–TAZ [pEF-TAZ-NFLAG, wild-type (wt) or the predominantly nuclear S89A mutant]. Cell extracts, antibodies [anti-HSV monoclonal antibody (αHSV, TEF-1) or the anti-HA antibody (αHA, Cntrl)] and Protein G–agarose were allowed to interact overnight at 4 °C. Proteins complexed to TEF-1ε–HSV were washed and separated by SDS/PAGE (10% gel). TAZ-N–FLAG (FL-TAZ) was identified by Western blot analysis with a polyclonal anti-YAP65/TAZ antibody. (B) YAP65 interacts with TEF-1. C2C12 cells were co-transfected with expression vectors encoding HA–YAP65 (pCMV-HA-YAP65) and FLAG–TEF-1ε (pCMV-FL-TEF-1ε). Complexes of FLAG–TEF-1 with HA–YAP65 were isolated by anti-FLAG–agarose (α-FL) or Protein G–agarose (pG) as control. Complexes were eluted from the FLAG–agarose with FLAG peptide, resolved by SDS/PAGE and HA–YAP65 (upper panel) and FLAG–TEF-1ε (FL-TEF-1; lower panel) were detected by Western blot with anti-HA (αHA) or anti-FLAG (αFLAG) antibodies. L, load. (C) Exogenous TEF-1 interacts with endogenous YAP65 and TAZ. H1299 cells were transfected with the FLAG–TEF-1 expression vector (pCMV-FL-TEF-1ε) and FLAG–TEF-1ε was isolated using anti-FLAG–agarose (α-FL) or Protein G–agarose (pG) as control. TEF-1 and associated proteins were separated by SDS/PAGE. Western blot analysis was performed with a polyclonal anti-YAP/TAZ antibody (αYAP/TAZ), showing that both endogenous TAZ and YAP65 are complexed to TEF-1 in vivo. Sizes in kDa are indicated in (B) and (C).
Figure 4. MCAT DNA-bound TEF-1 interacts with TAZ and YAP65
(A) Interaction of TEF-1 and TAZ on DNA was assayed by EMSAs. Purified TEF-1ε was mixed with [32P]MCAT DNA [MCAT1 from the cTNT promoter, containing a single MCAT site, or 2×GTIIC, containing two tandem MCAT sites separated by two base pairs (lanes 2 and 13)]. One (MCAT1) or two (2×GTIIC) TEF-1–MCAT complexes were formed. Increasing amounts of GST–TAZ-(1–239) were added to the reaction mixtures (lanes 2–7 and 13–18), causing formation of an LMC (TAZ–TEF-1–MCAT). Increasing TAZ concentrations caused the LMC to become more prominent. The LMC contains TEF-1 and TAZ, since both anti-GST (lanes 10 and 21) and anti-TEF-1 (lanes 11 and 22) antibodies supershifted this complex (Ab SS). No complexes were seen between MCAT DNA and GST alone (lanes 9 and 20) or between MCAT DNA and either the anti-GST or anti-TEF-1 antibodies (results not shown). (B) Interaction of TEF-1 and YAP65 on DNA was assayed by DNA-affinity chromatography. C2C12 cells were co-transfected with expression vectors encoding HA–YAP65 (pCMV-HA-YAP65) and FLAG–TEF-1ε (pCMV-FL-TEF-1ε). Complexes of FLAG–TEF-1ε with HA–YAP65 were isolated by and eluted from anti-FLAG–agarose as in Figure 3(B). Released complexes were purified further by MCAT DNA-affinity chromatography using biotinylated MCAT1 DNA [3]. After separation by SDS/PAGE, HA–YAP65 (upper panel) and FLAG–TEF-1 (lower panel) were detected by Western blot with anti-HA (αHA) or anti-FLAG (αFLAG) antibody. A significant portion of FLAG–TEF-1 was not purified by MCAT DNA (not bound). HA–YAP65 was found in fractions eluted from anti-FLAG–agarose (Load) and purified by MCAT DNA (Purif), indicating that YAP65 binds to TEF-1 on DNA. Sizes are indicated in kDa.
Figure 5. TAZ transactivates TEF-1-dependent transcription
(A) Mammalian one-hybrid assay. Expression vectors for GAL4 or GAL4–TEF-1-(1–426) (100 ng/six-well dish) and for FLAG–TAZ or HA–YAP65 (pEF-TAZ-NFAG and pCMV-HA-YAP65, 100 ng/well), a GAL4-luciferase reporter (pM5-luc, 250 ng) and Tk-Renilla (10 ng), as a co-transfection control, were co-transfected into H1299 cells using FuGENE 6. Extracts were made, and luciferase assays were performed as described in the Experimental section. Values (promoter activity) are expressed relative to the activity of the pM5-luc reporter in the presence of GAL4 and empty expression vector. TAZ and YAP65 interact with TEF-1, but not with GAL4 alone, to activate transcription. The human TEF-1 used to construct the GAL4 fusions used here is the equivalent of rat TEF-1ζ (426 amino acids, see Figure 2), which lacks a four-amino-acid alternatively spliced exon present in TEF-1ε (430 amino acids). (B) TAZ activates MCAT-dependent promoters. Luciferase assays were performed on H1299 cellular extracts from cells co-transfected with MCAT promoters (250 ng/dish) and TAZ expression vector (pEF-TAZ-NFLAG, 100 ng/dish). Values (fold activity) are presented relative to the −49(cTNT)-luc promoter in the presence of empty expression vector. Experiments were repeated four times, and results are means±S.E.M. All activations by TAZ were significant, P<0.05.
Figure 6. TAZ interacts with all four members of the TEF-1 family in vitro
(A) Each TEF-1 family member [TEF-1ε (T), RTEF-1 (R), DTEF-1 (D) and ETF-1 (E)] were produced and 35S-labelled by IVT reactions, and mixed with purified GST–TAZ-(1–239) (N), GST–TAZ-(239–395) (C) or GST (G). GST-fusion proteins and associated TEF-1 proteins were removed from solution with glutathione–agarose and assayed by SDS/PAGE. A total of 10% of each input is shown. Each member of the TEF-1 family specifically interacts with the N-terminal 239 amino acids of TAZ. TAZ interacts more efficiently with TEF-1ε (33% of input) than with RTEF-1 (22%). DTEF-1 and ETF-1 were produced much less efficiently than RTEF-1 and TEF-1ε (10–15%), so the images shown for these TEF-1 proteins are longer exposures, and the efficiency of interaction of DTEF-1 and ETF-1 with TAZ cannot be compared with that of TEF-1 or RTEF-1.
Figure 7. TAZ interacts more efficiently with TEF-1 than with RTEF-1
(A) TAZ binds TEF-1 more efficiently than RTEF-1 in vitro. GST pull-down assays were performed with GST–TAZ-(1–239) and 0–50 μl of IVT [35S]TEF-1ε or [35S]RTEF-1. The levels of input TEF-1s were quantified by trichloroacetic acid precipitation and phosphorimager analysis (see the Experimental section). The TAZ-bound TEF-1 and RTEF-1 were quantified by phosphorimager analysis and are expressed relative to the highest level of input 35S-labelled protein (approx. 1.3 pmol RTEF-1 at 50 μl of lysate). (B) Expression vectors for GAL4-fusion proteins [GAL4 DNA-binding domain alone, GAL4–TEF-1-(79–426), GAL4–RTEF-1-(79–426); 5, 10, 25, 50 and 100 ng], transcriptional co-activators (empty expression vector or pEF-TAZ-NFLAG, 100 ng), a GAL4–luciferase reporter (pM5-luc, 250 ng) and Tk-Renilla (10 ng), as a co-transfection control, were co-transfected into H1299 cells using FuGENE 6. Extracts were made, and luciferase assays were performed as described in the Experimental section. Promoter activity is expressed relative to the activity of the pM5-luc reporter in the presence of 5 ng of GAL4 and empty expression vector. TAZ was a more efficient co-activator for TEF-1 than for RTEF-1 at 5, 10 and 25 ng (P<0.05). If data are normalized to the basal activity of GAL4–TEF-1 and GAL4–RTEF-1, TAZ interacts more efficiently with TEF-1 than with RTEF-1 at all concentrations. Western blots to verify equal amounts of RTEF-1 were performed with plates transfected with the equivalent of 100 ng of GAL4 DNAs. Therefore equal production at low concentrations presumes a linear relationship between input DNA and protein production. Even if this assumption is not absolutely correct, since the activation levels of the reporter at 25 ng of GAL4–TEF-1 is approximately the same as for 10 ng of GAL4–RTEF-1, the conclusions of the experiment still hold, since it is unlikely that there is a 2.5-fold difference in protein levels.
Similar articles
- Vgl-4, a novel member of the vestigial-like family of transcription cofactors, regulates alpha1-adrenergic activation of gene expression in cardiac myocytes.
Chen HH, Mullett SJ, Stewart AF. Chen HH, et al. J Biol Chem. 2004 Jul 16;279(29):30800-6. doi: 10.1074/jbc.M400154200. Epub 2004 May 12. J Biol Chem. 2004. PMID: 15140898 - Hippo Component TAZ Functions as a Co-repressor and Negatively Regulates ΔNp63 Transcription through TEA Domain (TEAD) Transcription Factor.
Valencia-Sama I, Zhao Y, Lai D, Janse van Rensburg HJ, Hao Y, Yang X. Valencia-Sama I, et al. J Biol Chem. 2015 Jul 3;290(27):16906-17. doi: 10.1074/jbc.M115.642363. Epub 2015 May 20. J Biol Chem. 2015. PMID: 25995450 Free PMC article. - Effect of the acylation of TEAD4 on its interaction with co-activators YAP and TAZ.
Mesrouze Y, Meyerhofer M, Bokhovchuk F, Fontana P, Zimmermann C, Martin T, Delaunay C, Izaac A, Kallen J, Schmelzle T, Erdmann D, Chène P. Mesrouze Y, et al. Protein Sci. 2017 Dec;26(12):2399-2409. doi: 10.1002/pro.3312. Epub 2017 Nov 11. Protein Sci. 2017. PMID: 28960584 Free PMC article. - YAP/TAZ for cancer therapy: opportunities and challenges (review).
Guo L, Teng L. Guo L, et al. Int J Oncol. 2015 Apr;46(4):1444-52. doi: 10.3892/ijo.2015.2877. Epub 2015 Feb 5. Int J Oncol. 2015. PMID: 25652178 Review. - The Hippo Pathway and YAP/TAZ-TEAD Protein-Protein Interaction as Targets for Regenerative Medicine and Cancer Treatment.
Santucci M, Vignudelli T, Ferrari S, Mor M, Scalvini L, Bolognesi ML, Uliassi E, Costi MP. Santucci M, et al. J Med Chem. 2015 Jun 25;58(12):4857-73. doi: 10.1021/jm501615v. Epub 2015 Mar 11. J Med Chem. 2015. PMID: 25719868 Review.
Cited by
- A highly sensitive reporter system to monitor endogenous YAP1/TAZ activity and its application in various human cells.
Hikasa H, Kawahara K, Inui M, Yasuki Y, Yamashita K, Otsubo K, Kitajima S, Nishio M, Arima K, Endo M, Taira M, Suzuki A. Hikasa H, et al. Cancer Sci. 2024 Oct;115(10):3370-3383. doi: 10.1111/cas.16316. Epub 2024 Aug 18. Cancer Sci. 2024. PMID: 39155534 Free PMC article. - FAK, vinculin, and talin control mechanosensitive YAP nuclear localization.
Holland EN, Fernández-Yagüe MA, Zhou DW, O'Neill EB, Woodfolk AU, Mora-Boza A, Fu J, Schlaepfer DD, García AJ. Holland EN, et al. Biomaterials. 2024 Jul;308:122542. doi: 10.1016/j.biomaterials.2024.122542. Epub 2024 Mar 20. Biomaterials. 2024. PMID: 38547833 - Identification of a Gene Signature That Predicts Dependence upon YAP/TAZ-TEAD.
Kanai R, Norton E, Stern P, Hynes RO, Lamar JM. Kanai R, et al. Cancers (Basel). 2024 Feb 20;16(5):852. doi: 10.3390/cancers16050852. Cancers (Basel). 2024. PMID: 38473214 Free PMC article. - Keratinocyte integrin α3β1 induces expression of the macrophage stimulating factor, CSF-1, through a YAP/TEAD-dependent mechanism.
Longmate WM, Norton E, Duarte GA, Wu L, DiPersio MR, Lamar JM, DiPersio CM. Longmate WM, et al. Matrix Biol. 2024 Mar;127:48-56. doi: 10.1016/j.matbio.2024.02.003. Epub 2024 Feb 8. Matrix Biol. 2024. PMID: 38340968 - Current Model Systems for Investigating Epithelioid Haemangioendothelioma.
Neil E, Kouskoff V. Neil E, et al. Cancers (Basel). 2023 May 31;15(11):3005. doi: 10.3390/cancers15113005. Cancers (Basel). 2023. PMID: 37296967 Free PMC article. Review.
References
- Larkin S. B., Ordahl C. P. Multiple layers of control in transcriptional regulation by MCAT elements and the TEF-1 protein family. In: Harvey R. P., Rosenthal N., editors. Heart Development. San Diego: Academic Press; 1999. pp. 307–329.
- Farrance I. K., Mar J. H., Ordahl C. P. M-CAT binding factor is related to the SV40 enhancer binding factor, TEF-1. J. Biol. Chem. 1992;267:17234–17240. - PubMed
- Farrance I. K., Ordahl C. P. The role of transcription enhancer factor-1 (TEF-1) related proteins in the formation of M-CAT binding complexes in muscle and non-muscle tissues. J. Biol. Chem. 1996;271:8266–8274. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL27867/HL/NHLBI NIH HHS/United States
- R01 GM060954/GM/NIGMS NIH HHS/United States
- T32 AR07592/AR/NIAMS NIH HHS/United States
- T32 AR007592/AR/NIAMS NIH HHS/United States
- R01 HL071894/HL/NHLBI NIH HHS/United States
- GM60954/GM/NIGMS NIH HHS/United States
- HL071894/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous