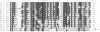Functional dissection of a conserved motif within the pilus retraction protein PilT - PubMed (original) (raw)
Functional dissection of a conserved motif within the pilus retraction protein PilT
Kelly G Aukema et al. J Bacteriol. 2005 Jan.
Abstract
PilT is a hexameric ATPase required for type IV pilus retraction in gram-negative bacteria. Retraction of type IV pili mediates intimate attachment to and signaling in host cells, surface motility, biofilm formation, natural transformation, and phage sensitivity. We investigated the in vivo and in vitro roles of each amino acid of the distinct, highly conserved C-terminal AIRNLIRE motif in PilT. Substitution of amino acids A288, I289, L292, and I293 as well as a double substitution of R290 and R294 abolished Pseudomonas aeruginosa PilT function in vivo, as measured by a loss of surface motility and phage sensitivity. When introduced into purified Aquifex aeolicus PilT, substitutions in the AIRNLIRE motif did not disrupt ATPase activity or oligomerization. In contrast, a K136Q substitution in the broadly conserved nucleotide binding motif prevented PilT function in vivo as well as in vitro. We propose that the AIRNLIRE motif forms an amphipathic alpha helix which transmits signals between a surface-exposed protein interaction site and the ATPase core of PilT, and we recognize a potential functional homology in other type II secretion ATPases.
Figures
FIG. 1.
Alignment of the C-terminal regions of in vivo functionally confirmed PilT and PilU proteins. The following sequences (NCBI reference number) are shown: Pa T, Pseudomonas aeruginosa PilT (AAG03784); Ps T, Pseudomonas stutzeri PilT (CAB56295); Rs T, Ralstonia solanacearum PilT (CAD16389); Ss T, Synechocystis sp. PilT (BAA18564); Nm T, Neisseria menigitidis PilT (AAF41181); Ng T, Neisseria gonorrhoeae PilT (AAB30824); Tt T, Thermus thermophilus PilT (AAL37755); Mx T, Myxococcus xanthus PilT (40); Aa T, Aquifex aeolicus PilT (AAC06903); Ps U, P. stutzeri PilU ((CAB56296); Pa U, P. aeruginosa PilU (S54702); Ng U, N. gonorrhoeae PilU (23); Ss T2, Synechocystis sp. PilT2 ((BAA18443); Pa B, P. aeruginosa PilB (AAG07914); and Vc E, Vibrio cholerae EpsE ((AAA58786). The alignment, created with Megalign Clustal W and manual optimization, begins with the last residues of the core nucleotide binding domain and the short glycine- and proline-rich linker region and is truncated at the end of the similarity, 6 to 20 amino acids from the C termini of the proteins. Residues that are identical or similar in 7 out of the 13 PilT and PilU sequences are shaded in black or grey, respectively; AIRNLIRE residues are indicated by asterisks. PilB and EpsE residues are shaded in black or grey if they are identical or similar, respectively, to conserved PilT and PilU residues. The amino acids of the tetracysteine loops of PilB and EpsE are not present in PilT or PilU. Percent identity (%I) was calculated from a pairwise alignment of each sequence with Pa T and from a pairwise alignment of Pa B with Vc E beginning at the caret (^).
FIG. 2.
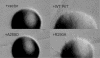
Colony morphology of cells harboring wild-type (WT) and mutated pilT genes. Representative colonies are shown for PAK ΔpilT plus the following plasmids: pUCP20 (+vector), pKA22 (+WT PilT), pKA56 (+A288D), and pKA60 (+R290A). Colonies are shown at the same magnifications.
FIG. 3.
Structural model of the proposed AIRNLIRE motif amphipathic α helix. Side-chain positions A288 and I293, at which single amino acid substitutions prevent both twitching motility and phage sensitivity in vivo (purple), suggest functional interactions. Additionally, the double mutant R290A/R294A loses PilT function. This stylized helix (grey carbon, red oxygen, and blue nitrogen atoms) was modeled in Xfit (16) by using standard side-chain conformations from the rotamer library and avoiding unfavorable steric clashes.
FIG. 4.
Accumulation of PilT proteins for all AIRNLIRE variants. Whole-cell lysates were examined by qualitative protein immunoblotting with an anti-gonococcal PilT antibody to visualize protein accumulation for plasmid-borne pilT variants of PAK ΔpilT. Each PilT variant is indicated by its amino acid substitution(s). Two gels are shown. Levels for I293 variants that were lower than those for the wild type were apparent and reproducible. Differences in accumulation for other variants were not reproducibly observed.
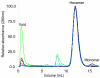
FIG. 5.
Size exclusion chromatography of purified wild-type and variant A. aeolicus PilT proteins. A total of 3 to 10 μg of each protein was injected. The PilT proteins are labeled as follows: A301D, red; I302A, pink; R303A, light green; N304A, light blue; I306A, black; R308A, brown; E308A, dark green; and wild type, dark blue. All chromatograms were normalized to a peak value of 100. The hexamer and monomer peaks are labeled (11). The apparent aggregation of R303A was not seen in all separations.
Similar articles
- Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU.
Chiang P, Sampaleanu LM, Ayers M, Pahuta M, Howell PL, Burrows LL. Chiang P, et al. Microbiology (Reading). 2008 Jan;154(Pt 1):114-126. doi: 10.1099/mic.0.2007/011320-0. Microbiology (Reading). 2008. PMID: 18174131 - Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility.
Satyshur KA, Worzalla GA, Meyer LS, Heiniger EK, Aukema KG, Misic AM, Forest KT. Satyshur KA, et al. Structure. 2007 Mar;15(3):363-76. doi: 10.1016/j.str.2007.01.018. Structure. 2007. PMID: 17355871 Free PMC article. - Cloning, expression, and functional analysis of molecular motor pilT and pilU genes of type IV pili in Acidithiobacillus ferrooxidans.
Li Y, Huang S, Zhang X, Huang T, Li H. Li Y, et al. Appl Microbiol Biotechnol. 2013 Feb;97(3):1251-7. doi: 10.1007/s00253-012-4271-1. Epub 2012 Jul 18. Appl Microbiol Biotechnol. 2013. PMID: 22805785 - The PIN-domain toxin-antitoxin array in mycobacteria.
Arcus VL, Rainey PB, Turner SJ. Arcus VL, et al. Trends Microbiol. 2005 Aug;13(8):360-5. doi: 10.1016/j.tim.2005.06.008. Trends Microbiol. 2005. PMID: 15993073 Review. - Pulling together with type IV pili.
Nudleman E, Kaiser D. Nudleman E, et al. J Mol Microbiol Biotechnol. 2004;7(1-2):52-62. doi: 10.1159/000077869. J Mol Microbiol Biotechnol. 2004. PMID: 15170403 Review.
Cited by
- Novel candidate virulence factors in rice pathogen Xanthomonas oryzae pv. oryzicola as revealed by mutational analysis.
Wang L, Makino S, Subedee A, Bogdanove AJ. Wang L, et al. Appl Environ Microbiol. 2007 Dec;73(24):8023-7. doi: 10.1128/AEM.01414-07. Epub 2007 Nov 2. Appl Environ Microbiol. 2007. PMID: 17981946 Free PMC article. - Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla.
Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ, Thomas BC, Banfield JF. Kantor RS, et al. mBio. 2013 Oct 22;4(5):e00708-13. doi: 10.1128/mBio.00708-13. mBio. 2013. PMID: 24149512 Free PMC article. - The type IV pilus protein PilU functions as a PilT-dependent retraction ATPase.
Adams DW, Pereira JM, Stoudmann C, Stutzmann S, Blokesch M. Adams DW, et al. PLoS Genet. 2019 Sep 16;15(9):e1008393. doi: 10.1371/journal.pgen.1008393. eCollection 2019 Sep. PLoS Genet. 2019. PMID: 31525185 Free PMC article. - Conformational changes in the motor ATPase CpaF facilitate a rotary mechanism of Tad pilus assembly.
Yen IY, Whitfield GB, Rubinstein JL, Burrows LL, Brun YV, Howell PL. Yen IY, et al. Nat Commun. 2025 Apr 24;16(1):3839. doi: 10.1038/s41467-025-59009-5. Nat Commun. 2025. PMID: 40268890 Free PMC article. - Change is good: variations in common biological mechanisms in the epsilonproteobacterial genera Campylobacter and Helicobacter.
Gilbreath JJ, Cody WL, Merrell DS, Hendrixson DR. Gilbreath JJ, et al. Microbiol Mol Biol Rev. 2011 Mar;75(1):84-132. doi: 10.1128/MMBR.00035-10. Microbiol Mol Biol Rev. 2011. PMID: 21372321 Free PMC article. Review.
References
- Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. - PubMed
- Bradley, D. E. 1972. Shortening of Pseudomonas aeruginosa pili after RNA-phage adsorption. J. Gen. Microbiol. 72:303-319. - PubMed
- Chou, P. Y., and G. D. Fasman. 1974. Prediction of protein conformation. Biochemistry 13:222-245. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases