The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli - PubMed (original) (raw)
The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli
Anna Z Nevesinjac et al. J Bacteriol. 2005 Jan.
Abstract
The Cpx envelope stress response mediates adaptation to potentially lethal envelope stresses in Escherichia coli. The two-component regulatory system consisting of the sensor kinase CpxA and the response regulator CpxR senses and mediates adaptation to envelope insults believed to result in protein misfolding in this compartment. Recently, a role was demonstrated for the Cpx response in the biogenesis of P pili, attachment organelles expressed by uropathogenic E. coli. CpxA senses misfolded P pilus assembly intermediates and initiates increased expression of both assembly and regulatory factors required for P pilus elaboration. In this report, we demonstrate that the Cpx response is also involved in the expression of the type IV bundle-forming pili of enteropathogenic E. coli (EPEC). Bundle-forming pili were not elaborated from an exogenous promoter in E. coli laboratory strain MC4100 unless the Cpx pathway was constitutively activated. Further, an EPEC cpxR mutant synthesized diminished levels of bundle-forming pili and was significantly affected in adherence to epithelial cells. Since type IV bundle-forming pili are very different from chaperone-usher-type P pili in both form and biogenesis, our results suggest that the Cpx envelope stress response plays a general role in the expression of envelope-localized organelles with diverse structures and assembly pathways.
Figures
FIG. 1.
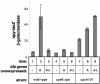
The Cpx pathway is activated by the overexpression of bfp genes. Expression from the spy promoter was measured by monitoring β-galactosidase expression from a _spy_-lacZ fusion harbored in MC4100 strains carrying control vector ptrc99A (−) (lanes 1, 4, and 7), plasmid pKDS301, which overexpresses bfpA (A) (lanes 2, 5, and 8), or plasmid pKDS302, which overexpresses the entire bfp gene cluster (A-L) (lanes 3, 6, and 9). Assays were performed with the wild-type strain (lanes 1 to 3), a cpxR1::Spc strain (lanes 4 to 6), and a strain carrying a cpxA101 allele (lanes 7 to 9). The strains used were as follows: lane 1, ALN17; lane 2, ALN18; lane 3, ALN19; lane 4, ALN33; lane 5, ALN34; lane 6, ALN35; lane 7, ALN40; lane 8, ALN41; and lane 9, ALN42. All assays were performed in triplicate and repeated at least twice. The data shown represent the mean and standard deviation from one experiment.
FIG. 2.
Constitutive activation of the Cpx response enables BFP expression in MC4100. BFP expression was examined in wild-type (WT) and cpxA* derivatives of MC4100 transformed with control vector ptrc99A (−), plasmid pKDS301, which overexpresses bfpA (A), or plasmid pKDS302, which overexpresses the entire bfp gene cluster (A-L). BFP expression was analyzed by Western blotting with antibundlin polyclonal antibody (A), by autoaggregation assays (B), and by transmission electron microscopy (C). The strains used were as follows: (A) lane 1, E2348/69; lane 2, ALN18; lane 3, ALN19; lane 4, ALN40; lane 5, ALN41; lane 6, ALN42; lane 7, ALN51; lane 8, ALN52; lane 9, ALN53; lane 10, ALN54; lane 11, ALN55; lane 12, ALN56; lane 13, ALN57; lane 14, ALN58; and lane 15, ALN59; (B) lane 1, ALN17; lane 2, ALN18; lane 3, ALN19; lane 4, ALN40; lane 5, ALN41; lane 6, ALN42; lane 7, ALN51; lane 8, ALN52; lane 9, ALN53; lane 10, ALN54; lane 11, ALN55; lane 12, ALN56; lane 13, ALN57; lane 14, ALN58; and lane 15, ALN59; and (C) ALN104. Western blotting was performed at least twice with independently isolated samples. Autoaggregation assays were performed in triplicate and repeated at least twice; data are reported as the mean and standard deviation. Electron microscopy was performed at least twice, and each time three separate grids were examined.
FIG. 2.
Constitutive activation of the Cpx response enables BFP expression in MC4100. BFP expression was examined in wild-type (WT) and cpxA* derivatives of MC4100 transformed with control vector ptrc99A (−), plasmid pKDS301, which overexpresses bfpA (A), or plasmid pKDS302, which overexpresses the entire bfp gene cluster (A-L). BFP expression was analyzed by Western blotting with antibundlin polyclonal antibody (A), by autoaggregation assays (B), and by transmission electron microscopy (C). The strains used were as follows: (A) lane 1, E2348/69; lane 2, ALN18; lane 3, ALN19; lane 4, ALN40; lane 5, ALN41; lane 6, ALN42; lane 7, ALN51; lane 8, ALN52; lane 9, ALN53; lane 10, ALN54; lane 11, ALN55; lane 12, ALN56; lane 13, ALN57; lane 14, ALN58; and lane 15, ALN59; (B) lane 1, ALN17; lane 2, ALN18; lane 3, ALN19; lane 4, ALN40; lane 5, ALN41; lane 6, ALN42; lane 7, ALN51; lane 8, ALN52; lane 9, ALN53; lane 10, ALN54; lane 11, ALN55; lane 12, ALN56; lane 13, ALN57; lane 14, ALN58; and lane 15, ALN59; and (C) ALN104. Western blotting was performed at least twice with independently isolated samples. Autoaggregation assays were performed in triplicate and repeated at least twice; data are reported as the mean and standard deviation. Electron microscopy was performed at least twice, and each time three separate grids were examined.
FIG. 2.
Constitutive activation of the Cpx response enables BFP expression in MC4100. BFP expression was examined in wild-type (WT) and cpxA* derivatives of MC4100 transformed with control vector ptrc99A (−), plasmid pKDS301, which overexpresses bfpA (A), or plasmid pKDS302, which overexpresses the entire bfp gene cluster (A-L). BFP expression was analyzed by Western blotting with antibundlin polyclonal antibody (A), by autoaggregation assays (B), and by transmission electron microscopy (C). The strains used were as follows: (A) lane 1, E2348/69; lane 2, ALN18; lane 3, ALN19; lane 4, ALN40; lane 5, ALN41; lane 6, ALN42; lane 7, ALN51; lane 8, ALN52; lane 9, ALN53; lane 10, ALN54; lane 11, ALN55; lane 12, ALN56; lane 13, ALN57; lane 14, ALN58; and lane 15, ALN59; (B) lane 1, ALN17; lane 2, ALN18; lane 3, ALN19; lane 4, ALN40; lane 5, ALN41; lane 6, ALN42; lane 7, ALN51; lane 8, ALN52; lane 9, ALN53; lane 10, ALN54; lane 11, ALN55; lane 12, ALN56; lane 13, ALN57; lane 14, ALN58; and lane 15, ALN59; and (C) ALN104. Western blotting was performed at least twice with independently isolated samples. Autoaggregation assays were performed in triplicate and repeated at least twice; data are reported as the mean and standard deviation. Electron microscopy was performed at least twice, and each time three separate grids were examined.
FIG. 3.
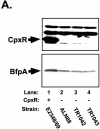
Mutation of cpxR in an EPEC strain background inhibits BFP production. CpxR and BFP levels were measured in a wild-type strain E2348/69 background (lanes 1) and in three independently isolated E2348/69 cpxR::Kn mutant strains, ALN88 (lanes 2), TR1042 (lanes 3), and TR1043 (lanes 4), by Western blotting (A and B) and autoaggregation assays (C) as described in Materials and Methods. Western blotting was performed multiple times, and one representative blot is shown. BfpA levels detected in panel A were quantitated with AlphaEase software and a FluorChem IS-5500 imaging system as described in Materials and Methods; the results are shown in panel B. BfpA levels were normalized to that of wild-type strain E2348/69, which was set at 100. Autoaggregation assays were performed in triplicate for each experiment and repeated at least twice; the data shown represent the mean and standard deviation from one experiment.
FIG. 3.
Mutation of cpxR in an EPEC strain background inhibits BFP production. CpxR and BFP levels were measured in a wild-type strain E2348/69 background (lanes 1) and in three independently isolated E2348/69 cpxR::Kn mutant strains, ALN88 (lanes 2), TR1042 (lanes 3), and TR1043 (lanes 4), by Western blotting (A and B) and autoaggregation assays (C) as described in Materials and Methods. Western blotting was performed multiple times, and one representative blot is shown. BfpA levels detected in panel A were quantitated with AlphaEase software and a FluorChem IS-5500 imaging system as described in Materials and Methods; the results are shown in panel B. BfpA levels were normalized to that of wild-type strain E2348/69, which was set at 100. Autoaggregation assays were performed in triplicate for each experiment and repeated at least twice; the data shown represent the mean and standard deviation from one experiment.
FIG. 3.
Mutation of cpxR in an EPEC strain background inhibits BFP production. CpxR and BFP levels were measured in a wild-type strain E2348/69 background (lanes 1) and in three independently isolated E2348/69 cpxR::Kn mutant strains, ALN88 (lanes 2), TR1042 (lanes 3), and TR1043 (lanes 4), by Western blotting (A and B) and autoaggregation assays (C) as described in Materials and Methods. Western blotting was performed multiple times, and one representative blot is shown. BfpA levels detected in panel A were quantitated with AlphaEase software and a FluorChem IS-5500 imaging system as described in Materials and Methods; the results are shown in panel B. BfpA levels were normalized to that of wild-type strain E2348/69, which was set at 100. Autoaggregation assays were performed in triplicate for each experiment and repeated at least twice; the data shown represent the mean and standard deviation from one experiment.
FIG. 4.
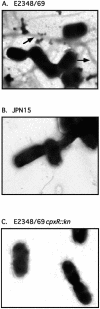
BFP are not detectable by transmission electron microscopy in EPEC _cpxR_-null mutants. BFP were examined by transmission electron microscopy of negatively stained samples of E2348/69 (A), JPN15 (B), and ALN88 (C) as described in Materials and Methods. Arrows indicate typical BFP seen. Samples were viewed at a magnification of ×14,000.
FIG. 5.
BfpA production is partially complemented in an E2348/69 cpxR::Kn background by a plasmid encoding cpxR. Levels of CpxR (A and B), bundlin (A and B), and surface-exposed BFP (C) were analyzed by Western blotting (A and B) and autoaggregation assays (C) of E2348/69 carrying control vector pBAD18 (lanes 1) or the E2348/69 cpxR::Kn mutant ALN88 transformed with pBAD18 (lanes 2) or CpxR overexpression plasmid pROX (lanes 3). Western blotting was performed at least twice. Levels of CpxR, BfpA, and a nonspecific band that cross-reacted with the CpxR antisera in panel A were quantitated with AlphaEase software and a FluorChem IS-5500 imaging system as described in Materials and Methods; the results are shown in panel B. Autoaggregation assays were performed in triplicate for each experiment and repeated at least twice; the data shown represent the mean and standard deviation from one experiment.
FIG. 5.
BfpA production is partially complemented in an E2348/69 cpxR::Kn background by a plasmid encoding cpxR. Levels of CpxR (A and B), bundlin (A and B), and surface-exposed BFP (C) were analyzed by Western blotting (A and B) and autoaggregation assays (C) of E2348/69 carrying control vector pBAD18 (lanes 1) or the E2348/69 cpxR::Kn mutant ALN88 transformed with pBAD18 (lanes 2) or CpxR overexpression plasmid pROX (lanes 3). Western blotting was performed at least twice. Levels of CpxR, BfpA, and a nonspecific band that cross-reacted with the CpxR antisera in panel A were quantitated with AlphaEase software and a FluorChem IS-5500 imaging system as described in Materials and Methods; the results are shown in panel B. Autoaggregation assays were performed in triplicate for each experiment and repeated at least twice; the data shown represent the mean and standard deviation from one experiment.
FIG. 5.
BfpA production is partially complemented in an E2348/69 cpxR::Kn background by a plasmid encoding cpxR. Levels of CpxR (A and B), bundlin (A and B), and surface-exposed BFP (C) were analyzed by Western blotting (A and B) and autoaggregation assays (C) of E2348/69 carrying control vector pBAD18 (lanes 1) or the E2348/69 cpxR::Kn mutant ALN88 transformed with pBAD18 (lanes 2) or CpxR overexpression plasmid pROX (lanes 3). Western blotting was performed at least twice. Levels of CpxR, BfpA, and a nonspecific band that cross-reacted with the CpxR antisera in panel A were quantitated with AlphaEase software and a FluorChem IS-5500 imaging system as described in Materials and Methods; the results are shown in panel B. Autoaggregation assays were performed in triplicate for each experiment and repeated at least twice; the data shown represent the mean and standard deviation from one experiment.
FIG. 6.
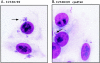
An EPEC cpxR mutant exhibits altered localized adherence. Localized-adherence assays were performed with E2348/69 (A) and ALN88 (B) as described in Materials and Methods. Arrows indicate typical clumps of adherent bacteria seen for each strain. The assays were performed in triplicate and repeated twice as described in Materials and Methods. The data represent typical fields of view. The statistical analysis is shown in Table 2.
Similar articles
- The Cpx envelope stress response both facilitates and inhibits elaboration of the enteropathogenic Escherichia coli bundle-forming pilus.
Vogt SL, Nevesinjac AZ, Humphries RM, Donnenberg MS, Armstrong GD, Raivio TL. Vogt SL, et al. Mol Microbiol. 2010 Jun 1;76(5):1095-110. doi: 10.1111/j.1365-2958.2010.07145.x. Epub 2010 Apr 14. Mol Microbiol. 2010. PMID: 20444097 Free PMC article. - Hfq reduces envelope stress by controlling expression of envelope-localized proteins and protein complexes in enteropathogenic Escherichia coli.
Vogt SL, Raivio TL. Vogt SL, et al. Mol Microbiol. 2014 May;92(4):681-97. doi: 10.1111/mmi.12581. Epub 2014 Apr 15. Mol Microbiol. 2014. PMID: 24628810 - Envelope stress responses and Gram-negative bacterial pathogenesis.
Raivio TL. Raivio TL. Mol Microbiol. 2005 Jun;56(5):1119-28. doi: 10.1111/j.1365-2958.2005.04625.x. Mol Microbiol. 2005. PMID: 15882407 Review. - Sensing external stress: watchdogs of the Escherichia coli cell envelope.
Ruiz N, Silhavy TJ. Ruiz N, et al. Curr Opin Microbiol. 2005 Apr;8(2):122-6. doi: 10.1016/j.mib.2005.02.013. Curr Opin Microbiol. 2005. PMID: 15802241 Review.
Cited by
- Revisiting long-chain fatty acid metabolism in Escherichia coli: integration with stress responses.
Jaswal K, Shrivastava M, Chaba R. Jaswal K, et al. Curr Genet. 2021 Aug;67(4):573-582. doi: 10.1007/s00294-021-01178-z. Epub 2021 Mar 19. Curr Genet. 2021. PMID: 33740112 Review. - A complex transcription network controls the early stages of biofilm development by Escherichia coli.
Prüss BM, Besemann C, Denton A, Wolfe AJ. Prüss BM, et al. J Bacteriol. 2006 Jun;188(11):3731-9. doi: 10.1128/JB.01780-05. J Bacteriol. 2006. PMID: 16707665 Free PMC article. Review. No abstract available. - Molecular insights into Escherichia coli Cpx envelope stress response activation by the sensor lipoprotein NlpE.
Marotta J, May KL, Bae CY, Grabowicz M. Marotta J, et al. Mol Microbiol. 2023 May;119(5):586-598. doi: 10.1111/mmi.15054. Epub 2023 Mar 25. Mol Microbiol. 2023. PMID: 36920223 Free PMC article. - Major Tom to Ground Control: How Lipoproteins Communicate Extracytoplasmic Stress to the Decision Center of the Cell.
Laloux G, Collet JF. Laloux G, et al. J Bacteriol. 2017 Oct 3;199(21):e00216-17. doi: 10.1128/JB.00216-17. Print 2017 Nov 1. J Bacteriol. 2017. PMID: 28674071 Free PMC article. Review. - The Cpx envelope stress response both facilitates and inhibits elaboration of the enteropathogenic Escherichia coli bundle-forming pilus.
Vogt SL, Nevesinjac AZ, Humphries RM, Donnenberg MS, Armstrong GD, Raivio TL. Vogt SL, et al. Mol Microbiol. 2010 Jun 1;76(5):1095-110. doi: 10.1111/j.1365-2958.2010.07145.x. Epub 2010 Apr 14. Mol Microbiol. 2010. PMID: 20444097 Free PMC article.
References
- Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources