Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors - PubMed (original) (raw)
. 2005 Jan 18;102(3):749-54.
doi: 10.1073/pnas.0408836102. Epub 2005 Jan 7.
Christina M Hughes, Ricardo Lloyd, Zhaohai Yang, Orit Rozenblatt-Rosen, Yali Dou, Robert W Schnepp, Cynthia Krankel, Virginia A Livolsi, Denise Gibbs, Xianxin Hua, Robert G Roeder, Matthew Meyerson, Jay L Hess
Affiliations
- PMID: 15640349
- PMCID: PMC545577
- DOI: 10.1073/pnas.0408836102
Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors
Thomas A Milne et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Mutations in the MEN1 gene are associated with the multiple endocrine neoplasia syndrome type 1 (MEN1), which is characterized by parathyroid hyperplasia and tumors of the pituitary and pancreatic islets. The mechanism by which MEN1 acts as a tumor suppressor is unclear. We have recently shown that menin, the MEN1 protein product, interacts with mixed lineage leukemia (MLL) family proteins in a histone methyltransferase complex including Ash2, Rbbp5, and WDR5. Here, we show that menin directly regulates expression of the cyclin-dependent kinase inhibitors p27Kip1 and p18Ink4c. Menin activates transcription by means of a mechanism involving recruitment of MLL to the p27Kip1 and p18Ink4c promoters and coding regions. Loss of function of either MLL or menin results in down-regulation of p27Kip1 and p18Ink4c expression and deregulated cell growth. These findings suggest that regulation of cyclin-dependent kinase inhibitor transcription by cooperative interaction between menin and MLL plays a central role in menin's activity as a tumor suppressor.
Figures
Fig. 1.
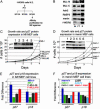
MLL and menin copurify in a complex that regulates expression of CDK inhibitors. (A) Strategy for immunoprecipitating MLL complex from HeLa cells expressing FLAG-tagged WDR-5. N.E., nuclear extract. (B) Western blots showing that menin and mammalian homologues of yeast SET1 complexes copurify with MLL. IN, input; FT, flow-through; E, eluate. (C) _Mll_-/- cells have a faster growth rate than Mll+/+ cells or three independent _Mll_-/- cells expressing FLAG-tagged MLL (F-MLL#1, F-MLL#6, and F-MLL#16). Growth curves were generated by plating cells at 1 × 105 in T75 flasks and counting cells at the same time point in successive 24-h periods by using Trypan blue exclusion. (Inset) Higher levels of p27Kip1 protein in Mll+/+ (lane 1), F-MLL#1 (lane 3), F-MLL#6 (lane 4), and F-MLL#16 (lane 5) compared with _Mll_-/- (lane 2) cell lines. (D) Menin-null cells (_Men1_-/-) and null cells with an empty expression vector (_Men1_-/- vec) have a faster growth rate than menin wild-type (Men1+/+) cells and null cells expressing exogenous menin (_Men1_-/- menin). (Inset) Higher p27Kip1 levels in slower Mll+/+ (lane 1), Men1+/+ (lane 3), and _Men1_-/- menin (5) compared with faster growing cells that are _Mll_-/- (lane 2), _Men1_-/- (lane 4), or _Men1_-/- cells with an empty expression vector (lane 6). Western blot for menin shows lack of expression in _Men1_-/- (lane 4) or _Men1_-/- cells with an empty expression vector (lane 6) and restoration of menin expression in _Men1_-/- menin cells (lane 5). (E) Real-Time PCR quantification using TaqMan probes shows higher expression of p27Kip1 and p18Ink4c transcripts in Mll+/+ (dark blue), F-MLL#6 (gray), and F-MLL#16 (red) cell lines compared with _Mll_-/- (green) cells. Gapdh and β-actin were both used as internal reference standards. TaqMan probe and primer sequences are available on request. (F) Quantitative RT-PCR shows higher expression of p27Kip1 and p18Ink4c transcripts in menin wild-type (Men1+/+, dark blue) or null cells with menin reexpression (_Men1_-/- men, red) compared with menin-null cells (_Men1_-/-, green) or menin-null cells harboring an empty expression vector (Men1_-/- vec, gray). Cell lines that express patient-derived menin point mutants (L22R, black; A242V, light blue) show defective restoration of p27Kip1 and p18Ink4c expression (Fig. 2_F Inset shows Western blot for menin expression in these cell lines).
Fig. 2.
Point mutations in menin interfere with binding and transcriptional regulation of CDK inhibitor loci. (A and B) Schematic of the p27Kip1 (A) and p18Ink4c (B) loci. Large arrow shows a major transcription start site at p27Kip1 conserved in both mice and humans. Smaller arrows show minor transcription start sites. Black bars are CpG rich regions; gray bars are exons. Red bar represents TaqMan primer/probe set. The initial ATG is indicated. (C and D) ChIP with an anti-menin antibody quantified with Real-Time PCR. TaqMan primer and probe sequences are available on request. (C) Menin binds to the p27 coding region in cells that are Mll+/+ (dark blue), _Mll_-/- (green), Men1+/+ (gray), or _Men1_-/- with reexpressed menin (light blue), but not in _Men1_-null cells or at the Gapdh locus in any cell type (_Men_-/-, red; _Men1_-/- with an empty vector, black). (D) Menin binds to the p18Ink4c coding region in cells that are Mll+/+ (dark blue), _Mll_-/- (green), Men1+/+ (gray), or _Men1_-/- with reexpressed menin (light blue). No binding is seen in _Men1_-null cells or at the Gapdh locus in any cell type (_Men1_-/-, red; _Men1_-/- with empty vector, black). (E) Luciferase assays on _Men1_-/- cells cotransfected with menin expression vector and p27Kip1 or p18Ink4c luciferase reporter shows menin activates transcription from the p27Kip1 or p18Ink4c promoters (green versus blue bars). The L22R (gray) and A242V (red) mutants show impaired ability to activate transcription. (F) Binding of the menin mutants L22R (gray) and A242V (red) are reduced at the p27Kip1 and p18Ink4c loci compared with wild-type menin (_Men1_-/- with reexpressed menin, green). (Inset) Western blot shows the L22R (gray lane 3) and A242V (red lane 4) mutants are expressed at similar levels to wild-type menin protein (green lane 2). Binding and menin expression in _Men1_-/- cells with an empty vector are shown as controls.
Fig. 3.
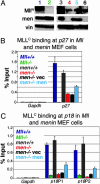
MLL binding to CDK inhibitor loci is reduced in the absence of menin. (A) Western blots for Mll and menin expression in cell lines used for ChIP. Although _Men_-/- cells (lane 4, red) express lower levels of Mll than Men1 wild-type cells (lane 3, gray), _Men_-/- cells with empty expression vector (lane 6, black) or menin expression vector (lane 5, light blue) show comparable levels of Mll expression. men, menin; vin, vinculin loading control. For comparison, Mll expression is also shown in Mll+/+ (lane 1, dark blue) and _Mll_-/- cells (lane 2, green). (B) ChIP with anti-MLLC antibody quantitated with Real-Time PCR shows that Mll binding to the p27Kip1 coding region is much higher in cells expressing menin. (C) Mll binding to the p18Ink4c coding region is also much higher in cells expressing menin. Binding to both p27Kip1 and p18Ink4c is high in Mll+/+ (dark blue), Men1+/+ (gray), or _Men1_-/- with menin reexpression (light blue) compared with _Men1_-null cells (_Men1_-/-, red), or _Men1_-/- with an empty expression vector (black). Negligible binding is detected in _Mll_-/- cells or at the Gapdh locus in any cell type.
Fig. 4.
MEN1 tumors show decreased p27Kip1 expression compared with normal endocrine tissues. (_A_-F) Immunoperoxidase staining for menin, MLL, and p27Kip1 in pancreatic tissues from a representative MEN1 patient. (A) Immunoperoxidase staining shows that menin is expressed in most morphologically normal islet cells. Expression is almost totally abolished in adenoma from the same tissue section (D). (B) MLL is expressed in essentially all normal islet cells. Expression is markedly decreased in adenoma from the same tissue section (E). (C) Immunoperoxidase staining for p27Kip1 shows high-level expression in 97% of islet cells (500 nuclei count). Expression is decreased both in number of cells (82%; 500 nuclei count) and intensity of staining in adenoma from the same tissue section(F). (G) Plot of percentage of p27Kip1-negative cells in normal islets and islet tumors based on 500 nuclei counts (n = 7). The blue horizontal bar represents average percent negative cells. (H) Model for role of MLL in normal homeostasis through regulation of Hox genes and CDK inhibitors (see text).
Similar articles
- Transcriptional activation of the mixed lineage leukemia-p27Kip1 pathway by a somatostatin analogue.
Horiguchi K, Yamada M, Satoh T, Hashimoto K, Hirato J, Tosaka M, Yamada S, Mori M. Horiguchi K, et al. Clin Cancer Res. 2009 Apr 15;15(8):2620-9. doi: 10.1158/1078-0432.CCR-08-2473. Epub 2009 Mar 24. Clin Cancer Res. 2009. PMID: 19318494 - Menin regulates pancreatic islet growth by promoting histone methylation and expression of genes encoding p27Kip1 and p18INK4c.
Karnik SK, Hughes CM, Gu X, Rozenblatt-Rosen O, McLean GW, Xiong Y, Meyerson M, Kim SK. Karnik SK, et al. Proc Natl Acad Sci U S A. 2005 Oct 11;102(41):14659-64. doi: 10.1073/pnas.0503484102. Epub 2005 Sep 29. Proc Natl Acad Sci U S A. 2005. PMID: 16195383 Free PMC article. - Haploinsufficient and predominant expression of multiple endocrine neoplasia type 1 (MEN1)-related genes, MLL, p27Kip1 and p18Ink4C in endocrine organs.
Taguchi R, Yamada M, Horiguchi K, Tomaru T, Ozawa A, Shibusawa N, Hashimoto K, Okada S, Satoh T, Mori M. Taguchi R, et al. Biochem Biophys Res Commun. 2011 Nov 18;415(2):378-83. doi: 10.1016/j.bbrc.2011.10.077. Epub 2011 Oct 21. Biochem Biophys Res Commun. 2011. PMID: 22037578 - Mechanisms of disease: multiple endocrine neoplasia type 1-relation to chromatin modifications and transcription regulation.
Dreijerink KM, Höppener JW, Timmers HM, Lips CJ. Dreijerink KM, et al. Nat Clin Pract Endocrinol Metab. 2006 Oct;2(10):562-70. doi: 10.1038/ncpendmet0292. Nat Clin Pract Endocrinol Metab. 2006. PMID: 17024155 Review. - Mechanisms of transformation by MLL.
Hess JL. Hess JL. Crit Rev Eukaryot Gene Expr. 2004;14(4):235-54. doi: 10.1615/critreveukaryotgeneexpr.v14.i4.10. Crit Rev Eukaryot Gene Expr. 2004. PMID: 15663355 Review.
Cited by
- Prognostic role of the CDNK1B V109G polymorphism in multiple endocrine neoplasia type 1.
Circelli L, Ramundo V, Marotta V, Sciammarella C, Marciello F, Del Prete M, Sabatino L, Pasquali D, Izzo F, Scala S, Colao A, Faggiano A, Colantuoni V; Multidisciplinary Group for NeuroEndocrine Tumours of Naples. Circelli L, et al. J Cell Mol Med. 2015 Jul;19(7):1735-41. doi: 10.1111/jcmm.12552. Epub 2015 Mar 30. J Cell Mol Med. 2015. PMID: 25824098 Free PMC article. - Restoring MLL reactivates latent tumor suppression-mediated vulnerability to proteasome inhibitors.
Ge M, Li D, Qiao Z, Sun Y, Kang T, Zhu S, Wang S, Xiao H, Zhao C, Shen S, Xu Z, Liu H. Ge M, et al. Oncogene. 2020 Sep;39(36):5888-5901. doi: 10.1038/s41388-020-01408-7. Epub 2020 Jul 30. Oncogene. 2020. PMID: 32733069 Free PMC article. - Epigenetic control of gene expression in leukemogenesis: Cooperation between wild type MLL and MLL fusion proteins.
Ballabio E, Milne TA. Ballabio E, et al. Mol Cell Oncol. 2014 Oct 29;1(2):e955330. doi: 10.1080/23723548.2014.955330. eCollection 2014 Apr-Jun. Mol Cell Oncol. 2014. PMID: 27308325 Free PMC article. Review. - RNA Sequencing and Pathway Analysis Identify Important Pathways Involved in Hypertrichosis and Intellectual Disability in Patients with Wiedemann-Steiner Syndrome.
Mietton L, Lebrun N, Giurgea I, Goldenberg A, Saintpierre B, Hamroune J, Afenjar A, Billuart P, Bienvenu T. Mietton L, et al. Neuromolecular Med. 2018 Sep;20(3):409-417. doi: 10.1007/s12017-018-8502-1. Epub 2018 Jul 16. Neuromolecular Med. 2018. PMID: 30014449 - Distribution of menin-occupied regions in chromatin specifies a broad role of menin in transcriptional regulation.
Agarwal SK, Impey S, McWeeney S, Scacheri PC, Collins FS, Goodman RH, Spiegel AM, Marx SJ. Agarwal SK, et al. Neoplasia. 2007 Feb;9(2):101-7. doi: 10.1593/neo.06706. Neoplasia. 2007. PMID: 17356705 Free PMC article.
References
- Chandrasekharappa, S. C., Guru, S. C., Manickam, P., Olufemi, S. E., Collins, F. S., Emmert-Buck, M. R., Debelenko, L. V., Zhuang, Z., Lubensky, I. A., Liotta, L. A., et al. (1997) Science 276, 404-407. - PubMed
- Lemmens, I., Van de Ven, W. J., Kas, K., Zhang, C. X., Giraud, S., Wautot, V., Buisson, N., De Witte, K., Salandre, J., Lenoir, G., et al. (1997) Hum. Mol. Genet. 6, 1177-1183. - PubMed
- Agarwal, S. K., Lee Burns, A., Sukhodolets, K. E., Kennedy, P. A., Obungu, V. H., Hickman, A. B., Mullendore, M. E., Whitten, I., Skarulis, M. C., Simonds, W. F., et al. (2004) Ann. N. Y. Acad. Sci. 1014, 189-198. - PubMed
- Bertolino, P., Tong, W. M., Herrera, P. L., Casse, H., Zhang, C. X. & Wang, Z. Q. (2003) Cancer Res. 63, 4836-4841. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous